Evolutionary artificial intelligence based peptide discoveries for effective Covid-19 therapeutics
- PMID: 32980462
- PMCID: PMC7832815
- DOI: 10.1016/j.bbadis.2020.165978
Evolutionary artificial intelligence based peptide discoveries for effective Covid-19 therapeutics
Abstract
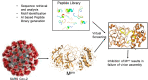
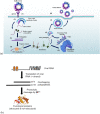
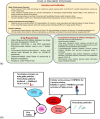
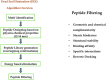
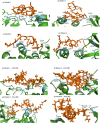
An epidemic caused by COVID-19 in China turned into pandemic within a short duration affecting countries worldwide. Researchers and companies around the world are working on all the possible strategies to develop a curative or preventive strategy for the same, which includes vaccine development, drug repurposing, plasma therapy, and drug discovery based on Artificial intelligence. Therapeutic approaches based on Computational biology and Machine-learning algorithms are specially considered, with a view that these could provide a fast and accurate outcome in the present scenario. As an effort towards developing possible therapeutics for COVID-19, we have used machine-learning algorithms for the generation of alignment kernels from diverse viral sequences of Covid-19 reported from India, China, Italy and USA. Using these diverse sequences we have identified the conserved motifs and subsequently a peptide library was designed against them. Of these, 4 peptides have shown strong binding affinity against the main protease of SARS-CoV-2 (Mpro) and also maintained their stability and specificity under physiological conditions as observed through MD Simulations. Our data suggest that these evolutionary peptides against COVID-19 if found effective may provide cross-protection against diverse Covid-19 variants.
Keywords: Artificial intelligence; Computational biology; Covid-19; Evolutionary peptides; SARS-CoV-2.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccines Immunother. 2020;16:1232–1238. doi: 10.1080/21645515.2020.1735227. - DOI - PMC - PubMed
-
- Wang Manli, Cao Ruiyuan, Zhang Leike, Yang Xinglou, Liu Jia, Xu Mingyue, Shi Zhengli, Hu Zhihong, Wu Zhong, Xiao Gengfu. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. https://www.nature.com/articles/s41422-020-0282-0 - PMC - PubMed
-
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta Mol. basis Dis. 1866;2020 doi: 10.1016/j.bbadis.2020.165878. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

