Long Non-Coding RNA LINC01089 Enhances the Development of Gastric Cancer by Sponging miR-145-5p to Mediate SOX9 Expression
- PMID: 32982308
- PMCID: PMC7508032
- DOI: 10.2147/OTT.S249392
Long Non-Coding RNA LINC01089 Enhances the Development of Gastric Cancer by Sponging miR-145-5p to Mediate SOX9 Expression
Retraction in
-
Long Non-Coding RNA LINC01089 Enhances the Development of Gastric Cancer by Sponging miR-145-5p to Mediate SOX9 Expression [Retraction].Onco Targets Ther. 2024 Jan 26;17:35-36. doi: 10.2147/OTT.S461192. eCollection 2024. Onco Targets Ther. 2024. PMID: 38292057 Free PMC article.
Abstract
Background: Long non-coding RNAs (lncRNAs) have potential regulatory effects in oncogenesis. Previous studies showed that several lncRNAs could participate in the progression of gastric cancer (GC). However, the specific biological mechanisms in GC are still unclear. We analyzed an lncRNA microarray of GC and selected LINC01089 for study.
Methods: LINC01089 expression in GC was tested by qRT-PCR. GC cell proliferation was assessed using CCK-8 and EdU assays. Cell invasion was assessed using the Transwell assay. A dual-luciferase reporter gene assay and bioinformatics assay were performed to detect potential targets of LINC01089. Additionally, RNA immunoprecipitation and Western blot assays were performed to clarify their interactions and roles in the regulation of GC progression.
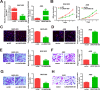
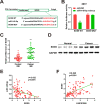
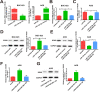
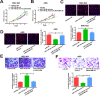
Results: High LINC01089 expression was observed in GC cells. LINC01089 overexpression notably expedited cell migration, proliferation, and invasion. LINC01089 positively regulated SOX9 expression by competitively binding to microRNA (miR-145-5p).
Conclusion: LINC01089 competitively binds to miR-145-5p to mediate SOX9 expression. LINC01089 may participate in the progression of GC.
Keywords: ceRNA; epigenetics; lncRNA; migration; proliferation.
© 2020 Wang and Yang.
Conflict of interest statement
The authors declare no competing interests for this work.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

