Exploiting Sphingo- and Glycerophospholipid Impairment to Select Effective Drugs and Biomarkers for CMT1A
- PMID: 32982928
- PMCID: PMC7477391
- DOI: 10.3389/fneur.2020.00903
Exploiting Sphingo- and Glycerophospholipid Impairment to Select Effective Drugs and Biomarkers for CMT1A
Abstract
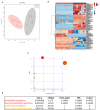
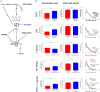
In Charcot-Marie-Tooth type 1A (CMT1A), Schwann cells exhibit a preponderant transcriptional deficiency of genes involved in lipid biosynthesis. This perturbed lipid metabolism affects the peripheral nerve physiology and the structure of peripheral myelin. Nevertheless, the identification and functional characterization of the lipid species mainly responsible for CMT1A myelin impairment currently lack. This is critical in the pathogenesis of the neuropathy since lipids are many and complex molecules which play essential roles in the cell, including the structural components of cellular membranes, cell signaling, and membrane trafficking. Moreover, lipids themselves are able to modify gene transcription, thereby affecting the genotype-phenotype correlation of well-defined inherited diseases, including CMT1A. Here we report for the first time a comprehensive lipid profiling in experimental and human CMT1A, demonstrating a previously unknown specific alteration of sphingolipid (SP) and glycerophospholipid (GP) metabolism. Notably, SP, and GP changes even emerge in biological fluids of CMT1A rat and human patients, implying a systemic metabolic dysfunction for these specific lipid classes. Actually, SP and GP are not merely reduced; their expression is instead aberrant, contributing to the ultrastructural abnormalities that we detailed by X-ray diffraction in rat and human internode myelin. The modulation of SP and GP pathways in myelinating dorsal root ganglia cultures clearly sustains this issue. In fact, just selected molecules interacting with these pathways are able to modify the altered geometric parameters of CMT1A myelinated fibers. Overall, we propose to exploit the present SP and GP metabolism impairment to select effective drugs and validate a set of reliable biomarkers, which remain a challenge in CMT1A neuropathy.
Keywords: CMT1A; Schwann cell; biomarker; demyelination; drug; lipid metabolism; myelin; peripheral neuropathy.
Copyright © 2020 Visigalli, Capodivento, Basit, Fernández, Hamid, Pencová, Gemelli, Marubbi, Pastorino, Luoma, Riekel, Kirschner, Schenone, Fernández, Armirotti and Nobbio.
Figures


References
LinkOut - more resources
Full Text Sources

