Rumen Microbiome Composition Is Altered in Sheep Divergent in Feed Efficiency
- PMID: 32983009
- PMCID: PMC7477290
- DOI: 10.3389/fmicb.2020.01981
Rumen Microbiome Composition Is Altered in Sheep Divergent in Feed Efficiency
Abstract
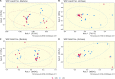
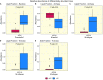
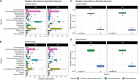
Rumen microbiome composition and functionality is linked to animal feed efficiency, particularly for bovine ruminants. To investigate this in sheep, we compared rumen bacterial and archaeal populations (and predicted metabolic processes) of sheep divergent for the feed efficiency trait feed conversion ratio (FCR). In our study 50 Texel cross Scottish Blackface (TXSB) ram lambs were selected from an original cohort of 200 lambs. From these, 26 were further selected for experimentation based on their extreme FCR (High Feed Efficiency, HFE = 13; Low Feed Efficiency, LFE = 13). Animals were fed a 95% concentrate diet ad libitum over 36 days. 16S rRNA amplicon sequencing was used to investigate the rumen bacterial and archaeal communities in the liquid and solid rumen fractions of sheep divergent for FCR. Weighted UniFrac distances separated HFE and LFE archaea communities from the liquid rumen fraction (Permanova, P < 0.05), with greater variation observed for the LFE cohort (Permdisp, P < 0.05). LFE animals exhibited greater Shannon and Simpson diversity indices, which was significant for the liquid rumen fraction (P < 0.05). Methanobrevibacter olleyae (in liquid and solid fractions) and Methanobrevibacter millerae (liquid fraction) were differentially abundant, and increased in the LFE cohort (P.adj < 0.05), while Methanobrevibacter wolinii (liquid fraction) was increased in the HFE cohort (P.adj < 0.05). This suggests that methanogenic archaea may be responsible for a potential loss of energy for the LFE cohort. Bacterial community composition (Permanova, P > 0.1) and diversity (P > 0.1) was not affected by the FCR phenotype. Only the genus Prevotella 1 was differentially abundant between HFE and LFE cohorts. Although no major compositional shifts of bacterial populations were identified amongst the feed efficient cohorts (FDR > 0.05), correlation analysis identified putative drivers of feed efficiency with Ruminococcaceae UCG-014 (liquid, rho = -0.53; solid, rho = -0.56) and Olsenella (solid, rho = -0.40) exhibiting significant negative association with FCR (P < 0.05). Bifidobacterium and Megasphaera showed significant positive correlations with ADG. Major cellulolytic bacteria Fibrobacter (liquid, rho = 0.43) and Ruminococcus 1 (liquid, rho = 0.41; solid, rho = 41) correlated positively with FCR (P < 0.05). Our study provides evidence that feed efficiency in sheep is likely influenced by compositional changes to the archaeal community, and abundance changes of specific bacteria, rather than major overall shifts within the rumen microbiome.
Keywords: 16S RNA; Greenhouse Gas; feed efficiency; metagenomics; microbiome; ruminant; sheep.
Copyright © 2020 McLoughlin, Spillane, Claffey, Smith, O’Rourke, Diskin and Waters.
Figures

Similar articles
-
Breed and ruminal fraction effects on bacterial and archaeal community composition in sheep.Sci Rep. 2023 Feb 27;13(1):3336. doi: 10.1038/s41598-023-28909-1. Sci Rep. 2023. PMID: 36849493 Free PMC article.
-
Metatranscriptomic Profiling Reveals Linkages between the Active Rumen Microbiome and Feed Efficiency in Beef Cattle.Appl Environ Microbiol. 2017 Apr 17;83(9):e00061-17. doi: 10.1128/AEM.00061-17. Print 2017 May 1. Appl Environ Microbiol. 2017. PMID: 28235871 Free PMC article.
-
Dietary wheat and reduced methane yield are linked to rumen microbiome changes in dairy cows.PLoS One. 2022 May 19;17(5):e0268157. doi: 10.1371/journal.pone.0268157. eCollection 2022. PLoS One. 2022. PMID: 35587477 Free PMC article.
-
Illumina MiSeq Phylogenetic Amplicon Sequencing Shows a Large Reduction of an Uncharacterised Succinivibrionaceae and an Increase of the Methanobrevibacter gottschalkii Clade in Feed Restricted Cattle.PLoS One. 2015 Jul 30;10(7):e0133234. doi: 10.1371/journal.pone.0133234. eCollection 2015. PLoS One. 2015. PMID: 26226343 Free PMC article.
-
Composition of bacterial and archaeal communities in the rumen of dromedary camel using cDNA-amplicon sequencing.Int Microbiol. 2020 May;23(2):137-148. doi: 10.1007/s10123-019-00093-1. Epub 2019 Aug 20. Int Microbiol. 2020. PMID: 31432356 Review.
Cited by
-
Dietary β-hydroxybutyric acid improves the growth performance of young ruminants based on rumen microbiota and volatile fatty acid biosynthesis.Front Microbiol. 2024 Jan 8;14:1296116. doi: 10.3389/fmicb.2023.1296116. eCollection 2023. Front Microbiol. 2024. PMID: 38260877 Free PMC article.
-
From Food Waste to Sustainable Agriculture: Nutritive Value of Potato By-Product in Total Mixed Ration for Angus Bulls.Foods. 2024 Aug 30;13(17):2771. doi: 10.3390/foods13172771. Foods. 2024. PMID: 39272536 Free PMC article.
-
Understanding Age-Related Longitudinal Dynamics in Abundance and Diversity of Dominant Culturable Gut Lactic Acid Bacteria in Pastured Goats.Animals (Basel). 2023 Aug 19;13(16):2669. doi: 10.3390/ani13162669. Animals (Basel). 2023. PMID: 37627460 Free PMC article.
-
Effects of different forage proportions in fermented total mixed ration on muscle fatty acid profile and rumen microbiota in lambs.Front Microbiol. 2023 Jul 13;14:1197059. doi: 10.3389/fmicb.2023.1197059. eCollection 2023. Front Microbiol. 2023. PMID: 37520349 Free PMC article.
-
Revealing the developmental characterization of rumen microbiome and its host in newly received cattle during receiving period contributes to formulating precise nutritional strategies.Microbiome. 2023 Nov 3;11(1):238. doi: 10.1186/s40168-023-01682-z. Microbiome. 2023. PMID: 37924150 Free PMC article.
References
-
- Abecia L., Martínez-Fernandez G., Waddams K., Martín-García A. I., Pinloche E., Creevey C. J., et al. (2018). Analysis of the rumen microbiome and metabolome to study the effect of an antimethanogenic treatment applied in early life of kid goats. Front. Microbiol. 9:2227. 10.3389/fmicb.2018.02227 - DOI - PMC - PubMed
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Cambridge: Babraham Institute.
LinkOut - more resources
Full Text Sources

