Toward Understanding Molecular Bases for Biological Diversification of Human Coronaviruses: Present Status and Future Perspectives
- PMID: 32983025
- PMCID: PMC7477919
- DOI: 10.3389/fmicb.2020.02016
Toward Understanding Molecular Bases for Biological Diversification of Human Coronaviruses: Present Status and Future Perspectives
Abstract
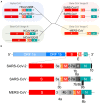
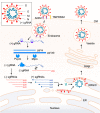
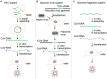
Human coronaviruses (HCoVs) are of zoonotic origins, and seven distinct HCoVs are currently known to infect humans. While the four seasonal HCoVs appear to be mildly pathogenic and circulate among human populations, the other three designated SARS-CoV, MERS-CoV, and SARS-CoV-2 can cause severe diseases in some cases. The newly identified SARS-CoV-2, a causative virus of COVID-19 that can be deadly, is now spreading worldwide much more efficiently than the other two pathogenic viruses. Despite evident differences in these properties, all HCoVs commonly have an exceptionally large genomic RNA with a rather peculiar gene organization and have the potential to readily alter their biological properties. CoVs are characterized by their biological diversifications, high recombination, and efficient adaptive evolution. We are particularly concerned about the high replication and transmission nature of SARS-CoV-2, which may lead to the emergence of more transmissible and/or pathogenic viruses than ever before. Furthermore, novel variant viruses may appear at any time from the CoV pools actively circulating or persistently being maintained in the animal reservoirs, and from the CoVs in infected human individuals. In this review, we describe knowns of the CoVs and then mention their unknowns to clarify the major issues to be addressed. Genome organizations and sequences of numerous CoVs have been determined, and the viruses are presently classified into separate phylogenetic groups. Functional roles in the viral replication cycle in vitro of non-structural and structural proteins are also quite well understood or suggested. In contrast, those in the in vitro and in vivo replication for various accessory proteins encoded by the variable 3' one-third portion of the CoV genome mostly remain to be determined. Importantly, the genomic sequences/structures closely linked to the high CoV recombination are poorly investigated and elucidated. Also, determinants for adaptation and pathogenicity have not been systematically investigated. We summarize here these research situations. Among conceivable projects, we are especially interested in the underlying molecular mechanism by which the observed CoV diversification is generated. Finally, as virologists, we discuss how we handle the present difficulties and propose possible research directions in the medium or long term.
Keywords: COVID-19; HCoV; MERS-CoV; SARS-CoV; SARS-CoV-2; adaptive evolution; biological diversification; recombination.
Copyright © 2020 Koma, Adachi, Doi, Adachi and Nomaguchi.
Figures
Similar articles
-
Zoonotic origins of human coronaviruses.Int J Biol Sci. 2020 Mar 15;16(10):1686-1697. doi: 10.7150/ijbs.45472. eCollection 2020. Int J Biol Sci. 2020. PMID: 32226286 Free PMC article. Review.
-
An overview on the seven pathogenic human coronaviruses.Rev Med Virol. 2022 Mar;32(2):e2282. doi: 10.1002/rmv.2282. Epub 2021 Aug 2. Rev Med Virol. 2022. PMID: 34339073 Review.
-
Antivirals for Broader Coverage against Human Coronaviruses.Viruses. 2024 Jan 20;16(1):156. doi: 10.3390/v16010156. Viruses. 2024. PMID: 38275966 Free PMC article. Review.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History.J Virol. 2017 Feb 14;91(5):e01953-16. doi: 10.1128/JVI.01953-16. Print 2017 Mar 1. J Virol. 2017. PMID: 28077633 Free PMC article.
Cited by
-
The role of airborne particles and environmental considerations in the transmission of SARS-CoV-2.Geosci Front. 2021 Sep;12(5):101189. doi: 10.1016/j.gsf.2021.101189. Epub 2021 Apr 5. Geosci Front. 2021. PMID: 38620834 Free PMC article. Review.
-
Variation and dispersal of PM10 and PM2.5 during COVID-19 lockdown over Kolkata metropolitan city, India investigated through HYSPLIT model.Geosci Front. 2022 Jan;13(1):101291. doi: 10.1016/j.gsf.2021.101291. Epub 2021 Aug 24. Geosci Front. 2022. PMID: 38620594 Free PMC article.
-
Genomic surveillance of SARS-CoV-2 tracks early interstate transmission of P.1 lineage and diversification within P.2 clade in Brazil.PLoS Negl Trop Dis. 2021 Oct 13;15(10):e0009835. doi: 10.1371/journal.pntd.0009835. eCollection 2021 Oct. PLoS Negl Trop Dis. 2021. PMID: 34644287 Free PMC article.
-
Current Status of Putative Animal Sources of SARS-CoV-2 Infection in Humans: Wildlife, Domestic Animals and Pets.Microorganisms. 2021 Apr 17;9(4):868. doi: 10.3390/microorganisms9040868. Microorganisms. 2021. PMID: 33920724 Free PMC article. Review.
-
The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions.Protein Sci. 2021 Jan;30(1):187-200. doi: 10.1002/pro.3978. Epub 2020 Nov 23. Protein Sci. 2021. PMID: 33070389 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

