Evaluation of the FilmArray® Pneumonia Plus Panel for Rapid Diagnosis of Hospital-Acquired Pneumonia in Intensive Care Unit Patients
- PMID: 32983057
- PMCID: PMC7477898
- DOI: 10.3389/fmicb.2020.02080
Evaluation of the FilmArray® Pneumonia Plus Panel for Rapid Diagnosis of Hospital-Acquired Pneumonia in Intensive Care Unit Patients
Abstract
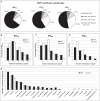
The FilmArray® Pneumonia plus Panel (FAPP) is a new multiplex molecular test for hospital-acquired pneumonia (HAP), which can rapidly detect 18 bacteria, 9 viruses, and 7 resistance genes. We aimed to compare the diagnosis performance of FAPP with conventional testing in 100 intensive care unit (ICU) patients who required mechanical ventilation, with clinically suspected HAP. A total of 237 samples [76 bronchoalveolar lavages (BALDS) and 82 endotracheal aspirates (ETADS) obtained at HAP diagnosis, and 79 ETA obtained during follow-up (ETATT)], were analyzed independently by routine microbiology testing and FAPP. 58 patients had paired BALDS and ETADS. The positivity thresholds of semi-quantified bacteria were 103-104 CFUs/mL or 104 copies/mL for BAL, and 105 CFUs/mL or copies/mL for ETA. Respiratory commensals (H. influenzae, S. aureus, E. coli, S. pneumoniae) were the most common pathogens. Discordant results for bacterial identification were observed in 33/76 (43.4%) BALDS and 36/82 (43.9%) ETADS, and in most cases, FAPP identified one supplemental bacteria (23/33 BALDS and 21/36 ETADS). An absence of growth, or polybacterial cultures, explained almost equally the majority of the non-detections in culture. No linear relationship was observed between bin and CFUs/mL variables. Concordant results between paired BALDS and ETADS were obtained in 46/58 (79.3%) patients with FAPP. One of the 17 resistance genes detected with FAPP (mecA/C and MREJ) was not confirmed by conventional testing. Overall, FAPP enhanced the positivity rate of diagnostic testing, with increased recognition of coinfections. Implementing this strategy may allow clinicians to make more timely and informed decisions.
Keywords: antibiotic resistance; coinfection; hospital-acquired pneumonia; multiplex syndromic testing; rapid diagnosis.
Copyright © 2020 Crémet, Gaborit, Bouras, Drumel, Guillotin, Poulain, Persyn, Lakhal, Rozec, Vibet, Roquilly and Gibaud.
Figures



References
-
- Buchan B. W., Windham S., Balada-Llasat J. M., Leber A., Harrington A., Relich R., et al. (2020). Practical comparison of the BioFire® FilmArray® Pneumonia Panel to routine diagnostic methods and potential impact on antimicrobial stewardship in adult hospitalized patients with lower respiratory tract infections. J. Clin. Microbiol. 29:JCM.00135-20. 10.1128/JCM.00135-20 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

