Single Nucleotide Polymorphism in KIR2DL1 Is Associated With HLA-C Expression in Global Populations
- PMID: 32983108
- PMCID: PMC7478174
- DOI: 10.3389/fimmu.2020.01881
Single Nucleotide Polymorphism in KIR2DL1 Is Associated With HLA-C Expression in Global Populations
Abstract
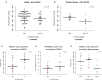
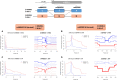
Regulation of NK cell activity is mediated through killer-cell immunoglobulin-like receptors (KIR) ability to recognize human leukocyte antigen (HLA) class I molecules as ligands. Interaction of KIR and HLA is implicated in viral infections, autoimmunity, and reproduction and there is growing evidence of the coevolution of these two independently segregating gene families. By leveraging KIR and HLA-C data from 1000 Genomes consortium we observed that the KIR2DL1 variant rs2304224*T is associated with lower expression of HLA-C in individuals carrying the ligand HLA-C2 (p = 0.0059). Using flow cytometry, we demonstrated that this variant is also associated with higher expression of KIR2DL1 on the NK cell surface (p = 0.0002). Next, we applied next generation sequencing to analyze KIR2DL1 sequence variation in 109 Euro and 75 Japanese descendants. Analyzing the extended haplotype homozygosity, we show signals of positive selection for rs4806553*G and rs687000*G, which are in linkage disequilibrium with rs2304224*T. Our results suggest that lower expression of HLA-C2 ligands might be compensated for higher expression of the receptor KIR2DL1 and bring new insights into the coevolution of KIR and HLA.
Keywords: KIR; NK cells; coevolution; expression; linkage disequilibrium; natural selection; population genetics.
Copyright © 2020 Vargas, Dourado, Amorim, Ho, Calonga-Solís, Issler, Marin, Beltrame, Petzl-Erler, Hollenbach and Augusto.
Figures
References
-
- van der Slik AR, Koeleman BPC, Verduijn W, Bruining GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. (2003) 52:2639–42. 10.2337/diabetes.52.10.2639 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

