Comparative Transcriptome Analysis of Litopenaeus vannamei Reveals That Triosephosphate Isomerase-Like Genes Play an Important Role During Decapod Iridescent Virus 1 Infection
- PMID: 32983114
- PMCID: PMC7485339
- DOI: 10.3389/fimmu.2020.01904
Comparative Transcriptome Analysis of Litopenaeus vannamei Reveals That Triosephosphate Isomerase-Like Genes Play an Important Role During Decapod Iridescent Virus 1 Infection
Abstract
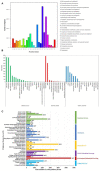
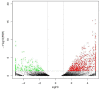
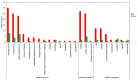
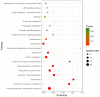
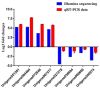
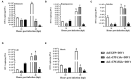
Decapod iridescent virus 1 (DIV1) results in severe economic losses in shrimp aquaculture. However, little is known about the physiological effect of DIV1 infection on the host. In this study, we found that the lethal dose 50 of DIV1-infected Litopenaeus vannamei after 48, 72, 96, and 156 h were 4.86 × 106, 5.07 × 105, 2.13 × 105, and 2.38 × 104 copies/μg DNA, respectively. In order to investigate the mechanisms of DIV1 infection, a comparative transcriptome analysis of hemocytes from L. vannamei, infected or not with DIV1, was conducted. The BUSCO analysis showed that the transcriptome was with high completeness (complete single-copy BUSCOs: 57.3%, complete duplicated BUSCOs: 41.1%, fragmentation: 0.8%, missing: 0.8%). A total of 168,854 unigenes were assembled, with an average length of 601 bp. Based on homology searches, Kyoto Encyclopedia of Genes and Genomes (KEGG), gene ontology (GO), and cluster of orthologous groups of proteins (KOG) analysis, 62,270 (36.88%) unigenes were annotated. Among them, 1,112 differentially expressed genes (DEGs) were identified, of which 889 genes were up-regulated and 223 genes were down-regulated after DIV1 infection. These genes were mainly annotated to the major metabolic processes such as fructose and mannose metabolism, carbon metabolism, and inositol phosphate metabolism. Among these metabolic pathways, the triosephosphate isomerase (TPI) family was the most eye-catching DEG as it participates in several metabolic processes. Three types of TPI, LvTPI-like, LvTPI-Blike, and LvTPI-Blike1, were obtained for gene silencing by RNA interference. The results showed that LvTPI-like and LvTPI-Blike1 silencing caused a high mortality rate among L. vannamei. However, LvTPI-like and LvTPI-Blike silencing reduced DIV1 replication in DIV1-infected L. vannamei. All the results indicated that TPI-like genes play an important role during DIV1 infection, which provides valuable insight into the infection mechanism of DIV1 in shrimp and may aid in preventing viral diseases in shrimp culture.
Keywords: DIV1; Litopenaeus vannamei; RNA interference; transcriptome analysis; triosephosphate isomerase.
Copyright © 2020 Liao, Wang, Wang, Qin, Hu, Wang, Sun and Zhang.
Figures




Similar articles
-
Integrated Analysis of mRNA-Seq and MiRNA-Seq Reveals the Molecular Mechanism of the Intestinal Immune Response in Marsupenaeus japonicus Under Decapod Iridescent Virus 1 Infection.Front Immunol. 2022 Jan 18;12:807093. doi: 10.3389/fimmu.2021.807093. eCollection 2021. Front Immunol. 2022. PMID: 35116034 Free PMC article.
-
Research into the hemocyte immune response of Fenneropenaeus merguiensis under decapod iridescent virus 1 (DIV1) challenge using transcriptome analysis.Fish Shellfish Immunol. 2020 Sep;104:8-17. doi: 10.1016/j.fsi.2020.05.053. Epub 2020 May 27. Fish Shellfish Immunol. 2020. PMID: 32473357
-
Transcriptomic analysis reveals the role of Glycolysis pathway in Litopenaeus vannamei during DIV1 infection.Fish Shellfish Immunol. 2023 Oct;141:109036. doi: 10.1016/j.fsi.2023.109036. Epub 2023 Aug 26. Fish Shellfish Immunol. 2023. PMID: 37640121
-
The Molecular Mechanism of Hemocyte Immune Response in Marsupenaeus japonicus Infected With Decapod Iridescent Virus 1.Front Microbiol. 2021 Aug 26;12:710845. doi: 10.3389/fmicb.2021.710845. eCollection 2021. Front Microbiol. 2021. PMID: 34512588 Free PMC article.
-
Infection with Decapod iridescent virus 1: an emerging disease in shrimp culture.Arch Microbiol. 2022 Nov 1;204(11):685. doi: 10.1007/s00203-022-03289-8. Arch Microbiol. 2022. PMID: 36319873 Review.
Cited by
-
Integrated analysis of intestinal microbiota and metabolomic reveals that decapod iridescent virus 1 (DIV1) infection induces secondary bacterial infection and metabolic reprogramming in Marsupenaeus japonicus.Front Immunol. 2022 Sep 16;13:982717. doi: 10.3389/fimmu.2022.982717. eCollection 2022. Front Immunol. 2022. PMID: 36189245 Free PMC article.
-
Integrated Analysis of mRNA-Seq and MiRNA-Seq Reveals the Molecular Mechanism of the Intestinal Immune Response in Marsupenaeus japonicus Under Decapod Iridescent Virus 1 Infection.Front Immunol. 2022 Jan 18;12:807093. doi: 10.3389/fimmu.2021.807093. eCollection 2021. Front Immunol. 2022. PMID: 35116034 Free PMC article.
-
GSK3β Plays a Negative Role During White Spot Syndrome Virus (WSSV) Infection by Regulating NF-κB Activity in Shrimp Litopenaeus vannamei.Front Immunol. 2020 Nov 26;11:607543. doi: 10.3389/fimmu.2020.607543. eCollection 2020. Front Immunol. 2020. PMID: 33324423 Free PMC article.
-
Host-microbiota interactions and responses of Metapenaeus ensis infected with decapod iridescent virus 1.Front Microbiol. 2023 Jan 13;13:1097931. doi: 10.3389/fmicb.2022.1097931. eCollection 2022. Front Microbiol. 2023. PMID: 36713173 Free PMC article.
-
A simple sequence repeats marker of disease resistance in shrimp Litopenaeus vannamei and its application in selective breeding.Front Genet. 2023 Jul 27;14:1144361. doi: 10.3389/fgene.2023.1144361. eCollection 2023. Front Genet. 2023. PMID: 37576558 Free PMC article.
References
-
- Thitamadee S, Prachumwat A, Srisala J, Jaroenlak P, Salachan PV, Sritunyalucksana K, et al. Review of current disease threats for cultivated penaeid shrimp in Asia. Aquaculture. (2016) 452:69–87. 10.1016/j.aquaculture.2015.10.028 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

