Epitranscriptomic N4-Acetylcytidine Profiling in CD4+ T Cells of Systemic Lupus Erythematosus
- PMID: 32984334
- PMCID: PMC7483482
- DOI: 10.3389/fcell.2020.00842
Epitranscriptomic N4-Acetylcytidine Profiling in CD4+ T Cells of Systemic Lupus Erythematosus
Abstract
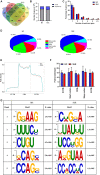
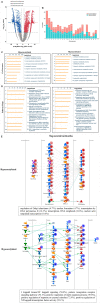
The emerging epitranscriptome plays an essential role in autoimmune disease. As a novel mRNA modification, N4-acetylcytidine (ac4C) could promote mRNA stability and translational efficiency. However, whether epigenetic mechanisms of RNA ac4C modification are involved in systemic lupus erythematosus (SLE) remains unclear. Herein, we detected eleven modifications in CD4+ T cells of SLE patients using mass spectrometry (LC-MS/MS). Furthermore, using samples from four CD4+ T cell pools, we identified lower modification of ac4C mRNA in SLE patients as compared to that in healthy controls (HCs). Meanwhile, significantly lower mRNA acetyltransferase NAT10 expression was detected in lupus CD4+ T cells by RT-qPCR. We then illustrated the transcriptome-wide ac4C profile in CD4+ T cells of SLE patients by ac4C-RIP-Seq and found ac4C distribution in mRNA transcripts to be highly conserved and enriched in mRNA coding sequence regions. Using bioinformatics analysis, the 3879 and 4073 ac4C hyper-acetylated and hypoacetylated peaks found in SLE samples, respectively, were found to be significantly involved in SLE-related function enrichments, including multiple metabolic and transcription-related processes, ROS-induced cellular signaling, apoptosis signaling, and NF-κB signaling. Moreover, we demonstrated the ac4C-modified regulatory network of gene biological functions in lupus CD4+ T cells. Notably, we determined that the 26 upregulated genes with hyperacetylation played essential roles in autoimmune diseases and disease-related processes. Additionally, the unique ac4C-related transcripts, including USP18, GPX1, and RGL1, regulate mRNA catabolic processes and translational initiation. Our study identified novel dysregulated ac4C mRNAs associated with critical immune and inflammatory responses, that have translational potential in lupus CD4+ T cells. Hence, our findings reveal transcriptional significance and potential therapeutic targets of mRNA ac4C modifications in SLE pathogenesis.
Keywords: CD4+ T cells; N4-acetylcytidine (ac4C); NAT10; epitranscriptome; systemic lupus erythematosus.
Copyright © 2020 Guo, Shi, Wang, Ye, Tong, Yan, Ding, Chen, Zhang and Xue.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

