Identification of Geographic Specific SARS-Cov-2 Mutations by Random Forest Classification and Variable Selection Methods
- PMID: 32984664
- PMCID: PMC7514111
Identification of Geographic Specific SARS-Cov-2 Mutations by Random Forest Classification and Variable Selection Methods
Abstract
RNA viral genomes have very high mutations rates. As infection spreads in the host populations, different viral lineages emerge acquiring independent mutations that can lead to varied infection and death rates in different parts of the world. By application of Random Forest classification and feature selection methods, we developed an analysis pipeline for identification of geographic specific mutations and classification of different viral lineages, focusing on the missense-variants that alter the function of the encoded proteins. We applied the pipeline on publicly available SARS-CoV-2 datasets and demonstrated that the analysis pipeline accurately identified country or region-specific viral lineages and specific mutations that discriminate different lineages. The results presented here can help designing country-specific diagnostic strategies and prioritizing the mutations for functional interpretation and experimental validations.
Keywords: Classification; Coronavirus; Feature selection; Random forest; SARS-CoV-2.
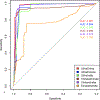
Figures


References
-
- Breiman L (1984). Classification and Regression Trees. Wadsworth International Group, Belmont, California.
-
- Breiman L (2001). Random Forests. Machine Learning 45, 5–32.
-
- Breiman L and Cutler A (2001). Random Forests. https://www.stat.berkeley.edu/~breiman/RandomForests/cc_home.htm.
-
- Cheng AS, Jin VX, Fan M, Smith LT et al. (2006). Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Molecular Cell, 21, 393–404. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
