Clinical features of COVID-19 mortality: development and validation of a clinical prediction model
- PMID: 32984797
- PMCID: PMC7508513
- DOI: 10.1016/S2589-7500(20)30217-X
Clinical features of COVID-19 mortality: development and validation of a clinical prediction model
Abstract
Background: The COVID-19 pandemic has affected millions of individuals and caused hundreds of thousands of deaths worldwide. Predicting mortality among patients with COVID-19 who present with a spectrum of complications is very difficult, hindering the prognostication and management of the disease. We aimed to develop an accurate prediction model of COVID-19 mortality using unbiased computational methods, and identify the clinical features most predictive of this outcome.
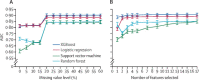
Methods: In this prediction model development and validation study, we applied machine learning techniques to clinical data from a large cohort of patients with COVID-19 treated at the Mount Sinai Health System in New York City, NY, USA, to predict mortality. We analysed patient-level data captured in the Mount Sinai Data Warehouse database for individuals with a confirmed diagnosis of COVID-19 who had a health system encounter between March 9 and April 6, 2020. For initial analyses, we used patient data from March 9 to April 5, and randomly assigned (80:20) the patients to the development dataset or test dataset 1 (retrospective). Patient data for those with encounters on April 6, 2020, were used in test dataset 2 (prospective). We designed prediction models based on clinical features and patient characteristics during health system encounters to predict mortality using the development dataset. We assessed the resultant models in terms of the area under the receiver operating characteristic curve (AUC) score in the test datasets.
Findings: Using the development dataset (n=3841) and a systematic machine learning framework, we developed a COVID-19 mortality prediction model that showed high accuracy (AUC=0·91) when applied to test datasets of retrospective (n=961) and prospective (n=249) patients. This model was based on three clinical features: patient's age, minimum oxygen saturation over the course of their medical encounter, and type of patient encounter (inpatient vs outpatient and telehealth visits).
Interpretation: An accurate and parsimonious COVID-19 mortality prediction model based on three features might have utility in clinical settings to guide the management and prognostication of patients affected by this disease. External validation of this prediction model in other populations is needed.
Funding: National Institutes of Health.
© 2020 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license.
Figures


Update of
-
Clinical predictors of COVID-19 mortality.medRxiv [Preprint]. 2020 May 22:2020.05.19.20103036. doi: 10.1101/2020.05.19.20103036. medRxiv. 2020. Update in: Lancet Digit Health. 2020 Oct;2(10):e516-e525. doi: 10.1016/S2589-7500(20)30217-X. PMID: 32511520 Free PMC article. Updated. Preprint.
Comment in
-
Prediction models for COVID-19 clinical decision making.Lancet Digit Health. 2020 Oct;2(10):e496-e497. doi: 10.1016/S2589-7500(20)30226-0. Epub 2020 Sep 22. Lancet Digit Health. 2020. PMID: 32984794 Free PMC article. No abstract available.
References
-
- Alpaydin E. 3rd edn. MIT Press; Cambridge, MA: 2014. Introduction to machine learning.
-
- US Centers for Disease Control and Prevention Health effects of second-hand smoke. Atlanta GA: US Centers for Disease Control and Prevention. https://www.cdc.gov/tobacco/data_statistics/fact_sheets/secondhand_smoke...
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

