An Optimized Protocol for In Vivo Analysis of Tumor Cell Division in a Sleeping Beauty-Mediated Mouse Glioma Model
- PMID: 32984853
- PMCID: PMC7518519
- DOI: 10.1016/j.xpro.2020.100044
An Optimized Protocol for In Vivo Analysis of Tumor Cell Division in a Sleeping Beauty-Mediated Mouse Glioma Model
Abstract
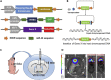
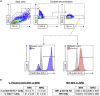
Malignant gliomas are the most common and aggressive primary brain tumor in adults, and high mitotic rates are associated with their malignancy. Gliomas were modeled in mice using the Sleeping Beauty system to encode genetic lesions recapitulating the human disease. The presented workflow allows the study of the proliferation of glioma cells in vivo, enabling the identification of different phases of the cell cycle, with the advantage that 5-ethynyl-2'-deoxyuridine staining does not involve denaturation steps and samples do not require histological processing. For complete details on the use and execution of this protocol, please refer to Núñez et al. (2019).
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures


References
-
- Brown D.V., Filiz G., Daniel P.M., Hollande F., Dworkin S., Amiridis S., Kountouri N., Ng W., Morokoff A.P., Mantamadiotis T. Expression of cd133 and cd44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS One. 2017;12:e0172791. - PMC - PubMed
-
- Garcia-Fabiani M.B., Comba A., Kadiyala P., Haase S., Núñez F.J., Altshuler D., Lowenstein P.R., Castro M.G. Isolation and characterization of immune cells from the tumor microenvironment of genetically engineered pediatric high-grade glioma models using the sleeping beauty transposon system. Methods Enzymol. 2020;632:369–388. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

