A Gold Nanoparticle Nanonuclease Relying on a Zn(II) Mononuclear Complex
- PMID: 32985766
- PMCID: PMC7839518
- DOI: 10.1002/anie.202012513
A Gold Nanoparticle Nanonuclease Relying on a Zn(II) Mononuclear Complex
Abstract
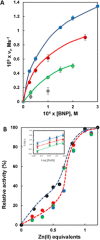
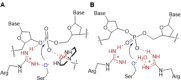
Similarly to enzymes, functionalized gold nanoparticles efficiently catalyze chemical reactions, hence the term nanozymes. Herein, we present our results showing how surface-passivated gold nanoparticles behave as synthetic nanonucleases, able to cleave pBR322 plasmid DNA with the highest efficiency reported so far for catalysts based on a single metal ion mechanism. Experimental and computational data indicate that we have been successful in creating a catalytic site precisely mimicking that suggested for natural metallonucleases relying on a single metal ion for their activity. It comprises one Zn(II) ion to which a phosphate diester of DNA is coordinated. Importantly, as in nucleic acids-processing enzymes, a positively charged arginine plays a key role by assisting with transition state stabilization and by reducing the pKa of the nucleophilic alcohol of a serine. Our results also show how designing a catalyst for a model substrate (bis-p-nitrophenylphosphate) may provide wrong indications as for its efficiency when it is tested against the real target (plasmid DNA).
Keywords: DNA cleavage; enzyme mimicry; nanonuclease; nanozymes; phosphate cleavage.
© 2020 The Authors. Published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Wolfenden R., Annu. Rev. Biochem. 2011, 80, 645–667. - PubMed
-
- Diez-Castellnou M., Martinez A., Mancin F., Adv. Phys. Org. Chem. 2017, 51, 129–186.
-
- Dupureur C., Curr. Opin. Chem. Biol. 2008, 12, 250–255. - PubMed
-
- Palermo G., Cavalli A., Klein M., Alfonso-Prieto M., Peraro M., De Vivo M., Acc. Chem. Res. 2015, 48, 220–228. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

