Optimized Transgene Delivery Using Third-Generation Lentiviruses
- PMID: 32986282
- PMCID: PMC7583475
- DOI: 10.1002/cpmb.125
Optimized Transgene Delivery Using Third-Generation Lentiviruses
Abstract
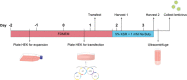
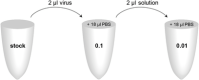
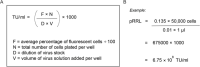
The lentivirus system enables efficient genetic modification of both dividing and non-dividing cells and therefore is a useful tool for elucidating developmental processes and disease pathogenesis. The development of third-generation lentiviruses has resulted in improved biosafety, low immunogenicity, and substantial packaging capabilities. However, because third-generation lentiviruses require successful co-transfection with four plasmids, this typically means that lower titers are attained. This is problematic, as it is often desirable to produce purified lentiviruses with high titers (>1 × 108 TU/ml), especially for in vivo applications. The manufacturing process for lentiviruses involves several critical experimental factors that can influence titer, purity, and transduction efficiency. Here, we describe a straightforward, stepwise protocol for the reproducible manufacture of high-titer third-generation lentiviruses (1 × 108 to 1 × 109 TU/ml). This optimized protocol enhances transgene expression by use of Lipofectamine transfection and optimized serum replacement medium, a single ultracentrifugation step, use of a sucrose cushion, and addition of a histone deacetylation inhibitor. Furthermore, we provide alternate methods for titration analyses, including functional and genomic integration analyses, using common laboratory techniques such as FACS as well as genomic DNA extraction and qPCR. These optimized methods will be beneficial for investigating developmental processes and disease pathogenesis in vitro and in vivo. © 2020 The Authors. Basic Protocol 1: Lentivirus production Support Protocol: Lentivirus concentration Basic Protocol 2: Lentivirus titration Alternate Protocol 1: Determination of viral titration by FACS analysis Alternate Protocol 2: Determination of viral titration by genome integration analysis.
Keywords: high titer; lentiviral production; lipofection; third-generation; ultracentrifugation.
© 2020 The Authors.
Figures


References
Literature Cited
-
- Boussif, O. , Lezoualc'h, F. , Zanta, M. A. , Mergny, M. D. , Scherman, D. , Demeneix, B. , & Behr, J. P. (1995). A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: Polyethylenimine. Proceedings of the National Academy of Sciences of the United States of America, 92(16), 7297–7301. doi: 10.1073/pnas.92.16.7297. - DOI - PMC - PubMed
-
- Burns, J. C. , Friedmann, T. , Driever, W. , Burrascano, M. , & Yee, J. K. (1993). Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: Concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 90(17), 8033–8037. doi: 10.1073/pnas.90.17.8033. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

