A bacterial size law revealed by a coarse-grained model of cell physiology
- PMID: 32986690
- PMCID: PMC7553314
- DOI: 10.1371/journal.pcbi.1008245
A bacterial size law revealed by a coarse-grained model of cell physiology
Abstract
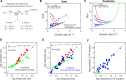
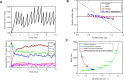
Universal observations in Biology are sometimes described as "laws". In E. coli, experimental studies performed over the past six decades have revealed major growth laws relating ribosomal mass fraction and cell size to the growth rate. Because they formalize complex emerging principles in biology, growth laws have been instrumental in shaping our understanding of bacterial physiology. Here, we discovered a novel size law that connects cell size to the inverse of the metabolic proteome mass fraction and the active fraction of ribosomes. We used a simple whole-cell coarse-grained model of cell physiology that combines the proteome allocation theory and the structural model of cell division. This integrated model captures all available experimental data connecting the cell proteome composition, ribosome activity, division size and growth rate in response to nutrient quality, antibiotic treatment and increased protein burden. Finally, a stochastic extension of the model explains non-trivial correlations observed in single cell experiments including the adder principle. This work provides a simple and robust theoretical framework for studying the fundamental principles of cell size determination in unicellular organisms.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Maaløe O. An analysis of bacterial growth. Dev Biol. 1969;3 (Suppl): 33–58.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

