Signatures of somatic mutations and gene expression from p16INK4A positive head and neck squamous cell carcinomas (HNSCC)
- PMID: 32986729
- PMCID: PMC7521680
- DOI: 10.1371/journal.pone.0238497
Signatures of somatic mutations and gene expression from p16INK4A positive head and neck squamous cell carcinomas (HNSCC)
Erratum in
-
Correction: Signatures of somatic mutations and gene expression from p16INK4A positive head and neck squamous cell carcinomas (HNSCC).PLoS One. 2024 Aug 8;19(8):e0308819. doi: 10.1371/journal.pone.0308819. eCollection 2024. PLoS One. 2024. PMID: 39116078 Free PMC article.
Abstract
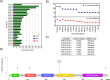
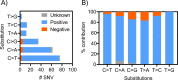
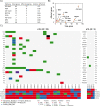
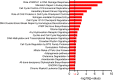
Human papilloma virus (HPV) causes a subset of head and neck squamous cell carcinomas (HNSCC) of the oropharynx. We combined targeted DNA- and genome-wide RNA-sequencing to identify genetic variants and gene expression signatures respectively from patients with HNSCC including oropharyngeal squamous cell carcinomas (OPSCC). DNA and RNA were purified from 35- formalin fixed and paraffin embedded (FFPE) HNSCC tumor samples. Immuno-histochemical evaluation of tumors was performed to determine the expression levels of p16INK4A and classified tumor samples either p16+ or p16-. Using ClearSeq Comprehensive Cancer panel, we examined the distribution of somatic mutations. Somatic single-nucleotide variants (SNV) were called using GATK-Mutect2 ("tumor-only" mode) approach. Using RNA-seq, we identified a catalog of 1,044 and 8 genes as significantly expressed between p16+ and p16-, respectively at FDR 0.05 (5%) and 0.1 (10%). The clinicopathological characteristics of the patients including anatomical site, smoking and survival were analyzed when comparing p16+ and p16- tumors. The majority of tumors (65%) were p16+. Population sequence variant databases, including gnomAD, ExAC, COSMIC and dbSNP, were used to identify the mutational landscape of somatic sequence variants within sequenced genes. Hierarchical clustering of The Cancer Genome Atlas (TCGA) samples based on HPV-status was observed using differentially expressed genes. Using RNA-seq in parallel with targeted DNA-seq, we identified mutational and gene expression signatures characteristic of p16+ and p16- HNSCC. Our gene signatures are consistent with previously published data including TCGA and support the need to further explore the biologic relevance of these alterations in HNSCC.
Conflict of interest statement
No Authors have competing interests.
Figures


References
-
- Saba NF, Goodman M, Ward K, Flowers C, Ramalingam S, Owonikoko T, et al. Gender and ethnic disparities in incidence and survival of squamous cell carcinoma of the oral tongue, base of tongue, and tonsils: a surveillance, epidemiology and end results program-based analysis. Oncology 2011;81(1):12–20 10.1159/000330807 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

