A comparative study of single nucleotide variant detection performance using three massively parallel sequencing methods
- PMID: 32986766
- PMCID: PMC7521702
- DOI: 10.1371/journal.pone.0239850
A comparative study of single nucleotide variant detection performance using three massively parallel sequencing methods
Abstract
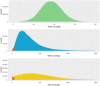
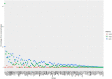
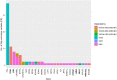
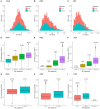
Massively parallel sequencing (MPS) has revolutionised clinical genetics and research within human genetics by enabling the detection of variants in multiple genes in several samples at the same time. Today, multiple approaches for MPS of DNA are available, including targeted gene sequencing (TGS) panels, whole exome sequencing (WES), and whole genome sequencing (WGS). As MPS is becoming an integrated part of the work in genetic laboratories, it is important to investigate the variant detection performance of the various MPS methods. We compared the results of single nucleotide variant (SNV) detection of three MPS methods: WGS, WES, and HaloPlex target enrichment sequencing (HES) using matched DNA of 10 individuals. The detection performance was investigated in 100 genes associated with cardiomyopathies and channelopathies. The results showed that WGS overall performed better than those of WES and HES. WGS had a more uniform and widespread coverage of the investigated regions compared to WES and HES, which both had a right-skewed coverage distribution and difficulties in covering regions and genes with high GC-content. WGS and WES showed roughly the same high sensitivities for detection of SNVs, whereas HES showed a lower sensitivity due to a higher number of false negative results.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Hertz CL, Christiansen SL, Ferrero-Miliani L, Dahl M, Weeke PE, LuCamp, et al. Next-generation sequencing of 100 candidate genes in young victims of suspected sudden cardiac death with structural abnormalities of the heart. Int J Legal Med. 2016;130(1):91–102. Epub 2015/09/19. 10.1007/s00414-015-1261-8 . - DOI - PubMed
-
- Nikiforova MN, Wald AI, Roy S, Durso MB, Nikiforov YE. Targeted Next-Generation Sequencing Panel (ThyroSeq) for Detection of Mutations in Thyroid Cancer. The Journal of Clinical Endocrinology & Metabolism. 2013;98(11):E1852–E60. 10.1210/jc.2013-2292 %J The Journal of Clinical Endocrinology & Metabolism. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

