A splice acceptor variant in HLA-DRA affects the conformation and cellular localization of the class II DR alpha-chain
- PMID: 32986852
- PMCID: PMC7808164
- DOI: 10.1111/imm.13273
A splice acceptor variant in HLA-DRA affects the conformation and cellular localization of the class II DR alpha-chain
Abstract
Class II human leucocyte antigen (HLA) proteins are involved in the immune response by presenting pathogen-derived peptides to CD4+ T lymphocytes. At the molecular level, they are constituted by α/β-heterodimers on the surface of professional antigen-presenting cells. Here, we report that the acceptor variant (rs8084) in the HLA-DRA gene mediates the transcription of an alternative version of the α-chain lacking 25 amino acids in its extracellular domain. Molecular dynamics simulations suggest this isoform undergoes structural refolding which in turn affects its stability and cellular trafficking. The short HLA-DRA isoform cannot reach the cell surface, although it is still able to bind the corresponding β-chain. Conversely, it remains entrapped within the endoplasmic reticulum where it is targeted for degradation. Furthermore, we demonstrate that the short isoform can be transported to the cell membrane via interactions with the peptide-binding site of canonical HLA heterodimers. Altogether, our findings indicate that short HLA-DRA functions as a novel intact antigen for class II HLA molecules.
Keywords: antigen presentation; human leucocyte antigen; immune response; protein folding.
© 2020 John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures

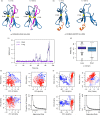

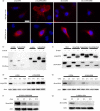


Similar articles
-
A short HLA-DRA isoform binds the HLA-DR2 heterodimer on the outer domain of the peptide-binding site.Arch Biochem Biophys. 2022 Apr 15;719:109156. doi: 10.1016/j.abb.2022.109156. Epub 2022 Feb 24. Arch Biochem Biophys. 2022. PMID: 35218721 Free PMC article.
-
DR alpha: E beta heterodimers in DRA transgenic mice hinder expression of E alpha: E beta molecules and are more efficient in antigen presentation.Int Immunol. 1995 Dec;7(12):1927-38. doi: 10.1093/intimm/7.12.1927. Int Immunol. 1995. PMID: 8746562
-
A mutant human histocompatibility leukocyte antigen DR molecule associated with invariant chain peptides.J Exp Med. 1994 Feb 1;179(2):541-9. doi: 10.1084/jem.179.2.541. J Exp Med. 1994. PMID: 8294865 Free PMC article.
-
How HLA-DM affects the peptide repertoire bound to HLA-DR molecules.Hum Immunol. 1997 May;54(2):170-9. doi: 10.1016/s0198-8859(97)00077-3. Hum Immunol. 1997. PMID: 9297535 Review.
-
The HLA system, antigen processing and presentation.Kidney Int Suppl. 1997 Mar;58:S2-7. Kidney Int Suppl. 1997. PMID: 9067934 Review.
Cited by
-
Screening and validation of differentially expressed genes in polymyositis.Heliyon. 2024 Jan 21;10(3):e24537. doi: 10.1016/j.heliyon.2024.e24537. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38317877 Free PMC article.
-
A short HLA-DRA isoform binds the HLA-DR2 heterodimer on the outer domain of the peptide-binding site.Arch Biochem Biophys. 2022 Apr 15;719:109156. doi: 10.1016/j.abb.2022.109156. Epub 2022 Feb 24. Arch Biochem Biophys. 2022. PMID: 35218721 Free PMC article.
-
A human leukocyte antigen imputation study uncovers possible genetic interplay between gut inflammatory processes and autism spectrum disorders.Transl Psychiatry. 2023 Jul 6;13(1):244. doi: 10.1038/s41398-023-02550-y. Transl Psychiatry. 2023. PMID: 37407551 Free PMC article.
References
-
- Kulski JK, Shiina T, Anzai T, Kohara S, Inoko H. Comparative genomic analysis of the MHC: the evolution of class I duplication blocks, diversity and complexity from shark to man. Immunol Rev. 2002; 190:95–122. - PubMed
-
- Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al Crystal structure of the human class II MHC protein HLA‐DR1 complexed with an influenza virus peptide. Nature. 1994; 368:215–21. - PubMed
-
- Runnels HA, Weber DA, Moore JC, Westerman LE, Jensen PE. Intact proteins can bind to class II histocompatibility molecules with high affinity. Mol Immunol. 1997; 34:471–80. - PubMed
-
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, et al Gene map of the extended human MHC. Nat Rev Genet. 2004; 5:889–99. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

