Design of novel viral attachment inhibitors of the spike glycoprotein (S) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) through virtual screening and dynamics
- PMID: 32987103
- PMCID: PMC7518233
- DOI: 10.1016/j.ijantimicag.2020.106177
Design of novel viral attachment inhibitors of the spike glycoprotein (S) of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) through virtual screening and dynamics
Abstract
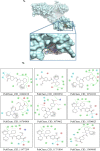
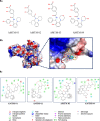
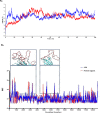
To date, the global COVID-19 pandemic has been associated with 11.8 million cases and over 545481 deaths. In this study, we have employed virtual screening approaches and selected 415 lead-like compounds from 103 million chemical substances, based on the existing drugs, from PubChem databases as potential candidates for the S protein-mediated viral attachment inhibition. Thereafter, based on drug-likeness and Lipinski's rules, 44 lead-like compounds were docked within the active side pocket of the viral-host attachment site of the S protein. Corresponding ligand properties and absorption, distribution, metabolism, excretion, and toxicity (ADMET) profile were measured. Furthermore, four novel inhibitors were designed and assessed computationally for efficacy. Comparative analysis showed the screened compounds in this study maintain better results than the proposed mother compounds, VE607 and SSAA09E2. The four designed novel lead compounds possessed more fascinating output without deviating from any of Lipinski's rules. They also showed higher bioavailability and the drug-likeness score was 0.56 and 1.81 compared with VE607 and SSAA09E2, respectively. All the screened compounds and novel compounds showed promising ADMET properties. Among them, the compound AMTM-02 was the best candidate, with a docking score of -7.5 kcal/mol. Furthermore, the binding study was verified by molecular dynamics simulation over 100 ns by assessing the stability of the complex. The proposed screened compounds and the novel compounds may give some breakthroughs for the development of a therapeutic drug to treat SARS-CoV-2 proficiently in vitro and in vivo.
Keywords: ADMET; Drug designing; Molecular docking; Molecular dynamics; Virtual screening.
Copyright © 2020 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of Competing Interests The authors declare no conflict of interest.
Figures






References
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. New Engl J Med. 2003;348(20):1953–1966. - PubMed
-
- Marra MA, Jones SJ, Astell CR, Holt RA, Brooks-Wilson A, Butterfield YS, et al. The genome sequence of the SARS-associated coronavirus. Science. 2003;300(5624):1399–1404. - PubMed
-
- Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300(5624):1394–1399. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

