The macro and micro of chromosome conformation capture
- PMID: 32987449
- PMCID: PMC8236208
- DOI: 10.1002/wdev.395
The macro and micro of chromosome conformation capture
Abstract
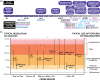
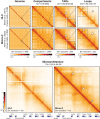
The 3D organization of the genome facilitates gene regulation, replication, and repair, making it a key feature of genomic function and one that remains to be properly understood. Over the past two decades, a variety of chromosome conformation capture (3C) methods have delineated genome folding from megabase-scale compartments and topologically associating domains (TADs) down to kilobase-scale enhancer-promoter interactions. Understanding the functional role of each layer of genome organization is a gateway to understanding cell state, development, and disease. Here, we discuss the evolution of 3C-based technologies for mapping 3D genome organization. We focus on genomics methods and provide a historical account of the development from 3C to Hi-C. We also discuss ChIP-based techniques that focus on 3D genome organization mediated by specific proteins, capture-based methods that focus on particular regions or regulatory elements, 3C-orthogonal methods that do not rely on restriction digestion and proximity ligation, and methods for mapping the DNA-RNA and RNA-RNA interactomes. We consider the biological discoveries that have come from these methods, examine the mechanistic contributions of CTCF, cohesin, and loop extrusion to genomic folding, and detail the 3D genome field's current understanding of nuclear architecture. Finally, we give special consideration to Micro-C as an emerging frontier in chromosome conformation capture and discuss recent Micro-C findings uncovering fine-scale chromatin organization in unprecedented detail. This article is categorized under: Gene Expression and Transcriptional Hierarchies > Regulatory Mechanisms Gene Expression and Transcriptional Hierarchies > Gene Networks and Genomics.
Keywords: 3C technologies; 3D genome; Hi-C; Micro-C; chromosome conformation capture; genomic organization; nuclear architecture.
© 2020 The Authors. WIREs Developmental Biology published by Wiley Periodicals LLC.
Conflict of interest statement
We declare that no competing financial interests exist.
Figures


References
-
- Andrey, G. , Schopflin, R. , Jerkovic, I. , Heinrich, V. , Ibrahim, D. M. , Paliou, C. , … Mundlos, S. (2017). Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Research, 27(2), 223–233. 10.1101/gr.213066.116 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

