A Rapid SARS-CoV-2 RT-PCR Assay for Low Resource Settings
- PMID: 32987722
- PMCID: PMC7598596
- DOI: 10.3390/diagnostics10100739
A Rapid SARS-CoV-2 RT-PCR Assay for Low Resource Settings
Abstract
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) assay is the gold standard recommended to test for acute SARS-CoV-2 infection. However, it generally requires expensive equipment such as RNA isolation instruments and real-time PCR thermal cyclers. As a pandemic, COVID-19 has spread indiscriminately, and many low resource settings and developing countries do not have the means for fast and accurate COVID-19 detection to control the outbreak. Additionally, long assay times, in part caused by slow sample preparation steps, have created a large backlog when testing patient samples suspected of COVID-19. With many PCR-based molecular assays including an extraction step, this can take a significant amount of time and labor, especially if the extraction is performed manually. Using COVID-19 clinical specimens, we have collected evidence that the RT-qPCR assay can feasibly be performed directly on patient sample material in virus transport medium (VTM) without an RNA extraction step, while still producing sensitive test results. If RNA extraction steps can be omitted without significantly affecting clinical sensitivity, the turn-around time of COVID-19 tests, and the backlog we currently experience can be reduced drastically. Furthermore, our data suggest that rapid RT-PCR can be implemented for sensitive and specific molecular diagnosis of COVID-19 in locations where sophisticated laboratory instruments are not available. Our USD 300 set up achieved rapid RT-PCR using thin-walled PCR tubes and a water bath setup using sous vide immersion heaters, a Raspberry Pi computer, and a single servo motor that can process up to 96 samples at a time. Using COVID-19 positive clinical specimens, we demonstrated that RT-PCR assays can be performed in as little as 12 min using untreated samples, heat-inactivated samples, or extracted RNA templates with our low-cost water bath setup. These findings can help rapid COVID-19 testing to become more accessible and attainable across the globe.
Keywords: RNA; SARS-CoV-2 virus; low resource setting; rapid RT-PCR assay; thermal cycler.
Conflict of interest statement
The authors declare no conflict of interest. A.A., P.Y., C.M. and M.W. are employed at AI Biosciences. S.W. is the co-founder of AI Biosciences. The other authors declare no competing financial interests.
Figures




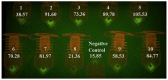
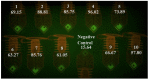
References
-
- Alcoba-Florez J., González-Montelongo R., Íñigo-Campos A., De Artola D.G.-M., Gil-Campesino H., Ciuffreda L., Valenzuela-Fernández A., Flores C., The Microbiology Technical Support Team Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. Int. J. Infect. Dis. 2020;97:66–68. doi: 10.1016/j.ijid.2020.05.099. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

