HSPA2 Chaperone Contributes to the Maintenance of Epithelial Phenotype of Human Bronchial Epithelial Cells but Has Non-Essential Role in Supporting Malignant Features of Non-Small Cell Lung Carcinoma, MCF7, and HeLa Cancer Cells
- PMID: 32987811
- PMCID: PMC7598654
- DOI: 10.3390/cancers12102749
HSPA2 Chaperone Contributes to the Maintenance of Epithelial Phenotype of Human Bronchial Epithelial Cells but Has Non-Essential Role in Supporting Malignant Features of Non-Small Cell Lung Carcinoma, MCF7, and HeLa Cancer Cells
Abstract
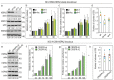
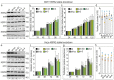
Heat Shock Protein A2 (HSPA2) is a member of the HSPA (HSP70) chaperone family and has a critical role for male fertility. HSPA2 is present in a number of somatic organs. Limited evidence suggests that HSPA2 may be involved in regulating epithelial cell differentiation. HSPA2 also emerged as a cancer-related chaperone; however, no consensus on its functional significance has been reached so far. In this study, we compared the phenotypic effects of HSPA2 deficit in non-transformed human bronchial epithelial cells (HBEC), and in lung, breast, and cervical cancer cells. We used various techniques to inhibit the HSPA2 gene expression in order to examine the impact of HSPA2 deficiency on cell growth, migration, adhesion, and invasion. Our results show that HBEC but not cancer cells are sensitive to HSPA2 deficit. HSPA2 knockdown in HBEC cells impaired their clone-forming ability and adhesiveness. Thus, our results indicate that epithelial cells can rely on a specific activity of HSPA2, but such dependence can be lost in epithelial cells that have undergone malignant transformation.
Keywords: HSP70; HSPA2; adhesion; breast cancer; bronchial epithelial cells; cell growth; cervical cancer; heat shock protein; malignant phenotype; non-small cell lung carcinoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








Similar articles
-
Novel role for the testis-enriched HSPA2 protein in regulating epidermal keratinocyte differentiation.J Cell Physiol. 2018 Mar;233(3):2629-2644. doi: 10.1002/jcp.26142. Epub 2017 Sep 7. J Cell Physiol. 2018. PMID: 28786487
-
Various Anti-HSPA2 Antibodies Yield Different Results in Studies on Cancer-Related Functions of Heat Shock Protein A2.Int J Mol Sci. 2020 Jun 16;21(12):4296. doi: 10.3390/ijms21124296. Int J Mol Sci. 2020. PMID: 32560263 Free PMC article.
-
Functional redundancy of HSPA1, HSPA2 and other HSPA proteins in non-small cell lung carcinoma (NSCLC); an implication for NSCLC treatment.Sci Rep. 2019 Oct 7;9(1):14394. doi: 10.1038/s41598-019-50840-7. Sci Rep. 2019. PMID: 31591429 Free PMC article.
-
Expression, function, and regulation of the testis-enriched heat shock HSPA2 gene in rodents and humans.Cell Stress Chaperones. 2015 Mar;20(2):221-35. doi: 10.1007/s12192-014-0548-x. Epub 2014 Oct 25. Cell Stress Chaperones. 2015. PMID: 25344376 Free PMC article. Review.
-
Heat Shock Protein A2 (HSPA2): Regulatory Roles in Germ Cell Development and Sperm Function.Adv Anat Embryol Cell Biol. 2017;222:67-93. doi: 10.1007/978-3-319-51409-3_4. Adv Anat Embryol Cell Biol. 2017. PMID: 28389751 Review.
Cited by
-
Identifying subtypes and developing prognostic models based on N6-methyladenosine and immune microenvironment related genes in breast cancer.Sci Rep. 2024 Jul 18;14(1):16586. doi: 10.1038/s41598-024-67477-w. Sci Rep. 2024. PMID: 39020010 Free PMC article.
-
Heat shock protein A2 is a novel extracellular vesicle-associated protein.Sci Rep. 2023 Mar 23;13(1):4734. doi: 10.1038/s41598-023-31962-5. Sci Rep. 2023. PMID: 36959387 Free PMC article.
-
Hub Genes PRPF19 and PPIB: Molecular Pathways and Potential Biomarkers in COPD.Int J Chron Obstruct Pulmon Dis. 2025 Jun 11;20:1865-1880. doi: 10.2147/COPD.S511696. eCollection 2025. Int J Chron Obstruct Pulmon Dis. 2025. PMID: 40524719 Free PMC article.
-
Inhibition of the Heat Shock Protein A (HSPA) Family Potentiates the Anticancer Effects of Manumycin A.Cells. 2021 Jun 7;10(6):1418. doi: 10.3390/cells10061418. Cells. 2021. PMID: 34200371 Free PMC article.
-
HSP70s in Breast Cancer: Promoters of Tumorigenesis and Potential Targets/Tools for Therapy.Cells. 2021 Dec 7;10(12):3446. doi: 10.3390/cells10123446. Cells. 2021. PMID: 34943954 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources

