Mechanisms of Deamidation of Asparagine Residues and Effects of Main-Chain Conformation on Activation Energy
- PMID: 32987875
- PMCID: PMC7582646
- DOI: 10.3390/ijms21197035
Mechanisms of Deamidation of Asparagine Residues and Effects of Main-Chain Conformation on Activation Energy
Abstract
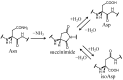
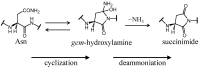
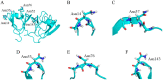
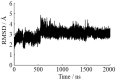
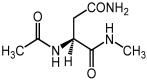
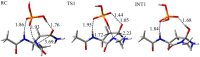
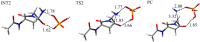
Deamidation of asparagine (Asn) residues is a nonenzymatic post-translational modification of proteins. Asn deamidation is associated with pathogenesis of age-related diseases and hypofunction of monoclonal antibodies. Deamidation rate is known to be affected by the residue following Asn on the carboxyl side and by secondary structure. Information about main-chain conformation of Asn residues is necessary to accurately predict deamidation rate. In this study, the effect of main-chain conformation of Asn residues on deamidation rate was computationally investigated using molecular dynamics (MD) simulations and quantum chemical calculations. The results of MD simulations for γS-crystallin suggested that frequently deamidated Asn residues have common main-chain conformations on the N-terminal side. Based on the simulated structure, initial structures for the quantum chemical calculations were constructed and optimized geometries were obtained using the B3LYP density functional method. Structures that were frequently deamidated had a lower activation energy barrier than that of the little deamidated structure. We also showed that dihydrogen phosphate and bicarbonate ions are important catalysts for deamidation of Asn residues.
Keywords: age-related diseases; deamidation; molecular dynamics simulation; post-translational modification; quantum chemical calculation.
Conflict of interest statement
The authors declare that they have no conflict of interest to disclose.
Figures
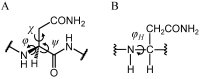


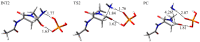

Similar articles
-
Investigation of the Effect of C-Terminal Adjacent Phenylalanine Residues on Asparagine Deamidation by Quantum Chemical Calculations.Int J Mol Sci. 2025 Jul 16;26(14):6819. doi: 10.3390/ijms26146819. Int J Mol Sci. 2025. PMID: 40725067 Free PMC article.
-
Identification of the Most Impactful Asparagine Residues for γS-Crystallin Aggregation by Deamidation.Biochemistry. 2023 Jun 6;62(11):1679-1688. doi: 10.1021/acs.biochem.3c00097. Epub 2023 May 8. Biochemistry. 2023. PMID: 37155656
-
Deamidation of Human γS-Crystallin Increases Attractive Protein Interactions: Implications for Cataract.Biochemistry. 2015 Aug 11;54(31):4890-9. doi: 10.1021/acs.biochem.5b00185. Epub 2015 Jul 29. Biochemistry. 2015. PMID: 26158710 Free PMC article.
-
Formulation considerations for proteins susceptible to asparagine deamidation and aspartate isomerization.J Pharm Sci. 2006 Nov;95(11):2321-36. doi: 10.1002/jps.20740. J Pharm Sci. 2006. PMID: 16960822 Review.
-
Mass spectrometric analysis of protein deamidation - A focus on top-down and middle-down mass spectrometry.Methods. 2022 Apr;200:58-66. doi: 10.1016/j.ymeth.2020.08.002. Epub 2020 Aug 10. Methods. 2022. PMID: 32791336 Review.
Cited by
-
General instability of dipeptides in concentrated sulfuric acid as relevant for the Venus cloud habitability.Sci Rep. 2024 Jul 24;14(1):17083. doi: 10.1038/s41598-024-67342-w. Sci Rep. 2024. PMID: 39048621 Free PMC article.
-
Designing Formulation Strategies for Enhanced Stability of Therapeutic Peptides in Aqueous Solutions: A Review.Pharmaceutics. 2023 Mar 14;15(3):935. doi: 10.3390/pharmaceutics15030935. Pharmaceutics. 2023. PMID: 36986796 Free PMC article. Review.
-
HLA-DQB1*05 subtypes and not DRB1*10:01 mediates risk in anti-IgLON5 disease.Brain. 2024 Jul 5;147(7):2579-2592. doi: 10.1093/brain/awae048. Brain. 2024. PMID: 38425314 Free PMC article.
-
Circular Dichroism Study of Orexin B under Oxidative Stress Conditions.Molecules. 2023 Jan 4;28(2):484. doi: 10.3390/molecules28020484. Molecules. 2023. PMID: 36677542 Free PMC article.
-
Investigation of the Effect of C-Terminal Adjacent Phenylalanine Residues on Asparagine Deamidation by Quantum Chemical Calculations.Int J Mol Sci. 2025 Jul 16;26(14):6819. doi: 10.3390/ijms26146819. Int J Mol Sci. 2025. PMID: 40725067 Free PMC article.
References
-
- Geiger T., Clarke S. Deamidation, isomerization, and racemization at asparaginyl and aspartyl residues in peptides. Succinimide-linked reactions that contribute to protein degradation. J. Biol. Chem. 1987;262:785–794. - PubMed
-
- Paranandi M.V., Guzzetta A.W., Hancock W.S., Aswad D.W. Deamidation and isoaspartate formation during in vitro aging of recombinant tissue plasminogen activator. J. Biol. Chem. 1994;269:243–253. - PubMed
-
- Robinson N.E., Robinson A.B. Molecular Clocks: Deamidation of Asparaginyl and Glutaminyl Residues in Peptides and Proteins. Althouse Press; Cave Junction, OR, USA: 2004. pp. 19–237.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

