Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies
- PMID: 32987909
- PMCID: PMC7599928
- DOI: 10.3390/v12101069
Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies
Abstract
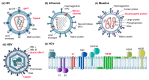
Structural virology reveals the architecture underlying infection. While notably electron microscopy images have provided an atomic view on viruses which profoundly changed our understanding of these assemblies incapable of independent life, spectroscopic techniques like NMR enter the field with their strengths in detailed conformational analysis and investigation of dynamic behavior. Typically, the large assemblies represented by viral particles fall in the regime of biological high-resolution solid-state NMR, able to follow with high sensitivity the path of the viral proteins through their interactions and maturation steps during the viral life cycle. We here trace the way from first solid-state NMR investigations to the state-of-the-art approaches currently developing, including applications focused on HIV, HBV, HCV and influenza, and an outlook to the possibilities opening in the coming years.
Keywords: capsids; membrane proteins; solid-state NMR; structure; viral proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

