Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula
- PMID: 32988416
- PMCID: PMC7523075
- DOI: 10.1186/s40168-020-00915-9
Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula
Erratum in
-
Correction to: Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula.Microbiome. 2021 May 10;9(1):105. doi: 10.1186/s40168-021-01080-3. Microbiome. 2021. PMID: 33971961 Free PMC article. No abstract available.
Abstract
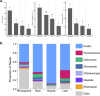
Background: Understanding the genetic and environmental factors that structure plant microbiomes is necessary for leveraging these interactions to address critical needs in agriculture, conservation, and sustainability. Legumes, which form root nodule symbioses with nitrogen-fixing rhizobia, have served as model plants for understanding the genetics and evolution of beneficial plant-microbe interactions for decades, and thus have added value as models of plant-microbiome interactions. Here we use a common garden experiment with 16S rRNA gene amplicon and shotgun metagenomic sequencing to study the drivers of microbiome diversity and composition in three genotypes of the model legume Medicago truncatula grown in two native soil communities.
Results: Bacterial diversity decreased between external (rhizosphere) and internal plant compartments (root endosphere, nodule endosphere, and leaf endosphere). Community composition was shaped by strong compartment × soil origin and compartment × plant genotype interactions, driven by significant soil origin effects in the rhizosphere and significant plant genotype effects in the root endosphere. Nevertheless, all compartments were dominated by Ensifer, the genus of rhizobia that forms root nodule symbiosis with M. truncatula, and additional shotgun metagenomic sequencing suggests that the nodulating Ensifer were not genetically distinguishable from those elsewhere in the plant. We also identify a handful of OTUs that are common in nodule tissues, which are likely colonized from the root endosphere.
Conclusions: Our results demonstrate strong host filtering effects, with rhizospheres driven by soil origin and internal plant compartments driven by host genetics, and identify several key nodule-inhabiting taxa that coexist with rhizobia in the native range. Our results set the stage for future functional genetic experiments aimed at expanding our pairwise understanding of legume-rhizobium symbiosis toward a more mechanistic understanding of plant microbiomes. Video Abstract.
Keywords: Common garden; Evolution; Genetic variation; Mutualism; Nodule microbiome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Rhizobial nitrogen fixation efficiency shapes endosphere bacterial communities and Medicago truncatula host growth.Microbiome. 2023 Jul 3;11(1):146. doi: 10.1186/s40168-023-01592-0. Microbiome. 2023. PMID: 37394496 Free PMC article.
-
The Influence of the Host Plant Is the Major Ecological Determinant of the Presence of Nitrogen-Fixing Root Nodule Symbiont Cluster II Frankia Species in Soil.Appl Environ Microbiol. 2016 Dec 15;83(1):e02661-16. doi: 10.1128/AEM.02661-16. Print 2017 Jan 1. Appl Environ Microbiol. 2016. PMID: 27795313 Free PMC article.
-
Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments.Mol Ecol. 2017 Mar;26(6):1641-1651. doi: 10.1111/mec.14027. Epub 2017 Feb 23. Mol Ecol. 2017. PMID: 28139080
-
Medicago truncatula proteomics.J Proteomics. 2010 Sep 10;73(10):1974-85. doi: 10.1016/j.jprot.2010.07.004. Epub 2010 Jul 16. J Proteomics. 2010. PMID: 20621211 Review.
-
The Role of Host Genetic Signatures on Root-Microbe Interactions in the Rhizosphere and Endosphere.Front Plant Sci. 2018 Dec 19;9:1896. doi: 10.3389/fpls.2018.01896. eCollection 2018. Front Plant Sci. 2018. PMID: 30619438 Free PMC article. Review.
Cited by
-
Rational management of the plant microbiome for the Second Green Revolution.Plant Commun. 2024 Apr 8;5(4):100812. doi: 10.1016/j.xplc.2024.100812. Epub 2024 Jan 11. Plant Commun. 2024. PMID: 38213028 Free PMC article. Review.
-
In vitro antioxidant and antibacterial activities of Ajuga integrifolia leaf extracts obtained with different solvents.BMC Complement Med Ther. 2024 Oct 12;24(1):368. doi: 10.1186/s12906-024-04668-y. BMC Complement Med Ther. 2024. PMID: 39395997 Free PMC article.
-
Patterns in the Microbial Community of Salt-Tolerant Plants and the Functional Genes Associated with Salt Stress Alleviation.Microbiol Spectr. 2021 Oct 31;9(2):e0076721. doi: 10.1128/Spectrum.00767-21. Epub 2021 Oct 27. Microbiol Spectr. 2021. PMID: 34704793 Free PMC article.
-
Compartment Niche Shapes the Assembly and Network of Cannabis sativa-Associated Microbiome.Front Microbiol. 2021 Oct 5;12:714993. doi: 10.3389/fmicb.2021.714993. eCollection 2021. Front Microbiol. 2021. PMID: 34675893 Free PMC article.
-
Long-distance movement dynamics shape host microbiome richness and turnover.FEMS Microbiol Ecol. 2024 Jun 17;100(7):fiae089. doi: 10.1093/femsec/fiae089. FEMS Microbiol Ecol. 2024. PMID: 38857884 Free PMC article.
References
-
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. - PubMed
-
- Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Métraux J-P, L’Haridon F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016;210:1033–1043. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

