Transcriptomic and Proteomic Insights into Amborella trichopoda Male Gametophyte Functions
- PMID: 32989009
- PMCID: PMC7723084
- DOI: 10.1104/pp.20.00837
Transcriptomic and Proteomic Insights into Amborella trichopoda Male Gametophyte Functions
Abstract
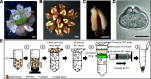
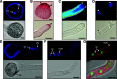
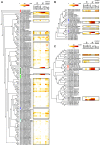
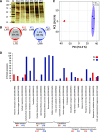
Flowering plants (angiosperms) are characterized by pollen tubes (PTs; male gametophytes) carrying two immobile sperm cells that grow over long distances through the carpel toward the ovules, where double fertilization is executed. It is not understood how these reproductive structures evolved, which genes occur de novo in male gametophytes of angiosperms, and to which extent PT functions are conserved among angiosperms. To contribute to a deeper understanding of the evolution of gametophyte functions, we generated RNA sequencing data from seven reproductive and two vegetative control tissues of the basal angiosperm Amborella trichopoda and complemented these with proteomic data of pollen grains (PGs) and PTs. The eudicot model plant Arabidopsis (Arabidopsis thaliana) served as a reference organism for data analysis, as more than 200 genes have been associated with male gametophyte functions in this species. We describe methods to collect bicellular A. trichopoda PGs, to induce their germination in vitro, and to monitor PT growth and germ cell division. Transcriptomic and proteomic analyses indicate that A. trichopoda PGs are prepared for germination requiring lipids, energy, but likely also reactive oxygen species, while PTs are especially characterized by catabolic/biosynthetic and transport processes including cell wall biosynthesis and gene regulation. Notably, a number of pollen-specific genes were lacking in Arabidopsis, and the number of genes involved in pollen signaling is significantly reduced in A. trichopoda In conclusion, we provide insight into male gametophyte functions of the most basal angiosperm and establish a valuable resource for future studies on the evolution of flowering plants.
© 2020 American Society of Plant Biologists. All Rights Reserved.
Figures



References
-
- Amborella Genome Project (2013) The Amborella genome and the evolution of flowering plants. Science 342: 1241089. - PubMed
-
- Benjamini Y, Hochberg Y(1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

