Transcriptional Responses of Pseudomonas aeruginosa to Inhibition of Lipoprotein Transport by a Small Molecule Inhibitor
- PMID: 32989085
- PMCID: PMC7685553
- DOI: 10.1128/JB.00452-20
Transcriptional Responses of Pseudomonas aeruginosa to Inhibition of Lipoprotein Transport by a Small Molecule Inhibitor
Abstract
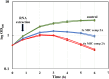
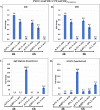
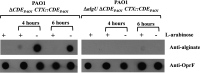
Lipoprotein transport from the inner to the outer membrane, carried out by the Lol machinery, is essential for the biogenesis of the Gram-negative cell envelope and, consequently, for bacterial viability. Recently, small molecule inhibitors of the Lol system in Escherichia coli have been identified and shown to inhibit the growth of this organism by interfering with the function of the LolCDE complex. Analysis of the transcriptome of E. coli treated with one such molecule (compound 2) revealed that a number of envelope stress response pathways were induced in response to LolCDE inhibition. However, Pseudomonas aeruginosa is refractory to inhibition by the same small molecule, but we could demonstrate that E. colilolCDE could be substituted for the P. aeruginosa orthologues, where it functions in the correct transport of Pseudomonas lipoproteins, and the cells are inhibited by the more potent compound 2A. In the present study, we took advantage of the functionality of E. coli LolCDE in P. aeruginosa and determined the P. aeruginosa transcriptional response to LolCDE inhibition by compound 2A. We identified key genes that responded to LolCDE inhibition and also demonstrated that the same genes appeared to be affected by genetic depletion of the native P. aeruginosa LolCDE proteins. Several of the major changes were in an upregulated cluster of genes that encode determinants of alginate biosynthesis and transport, and the levels of alginate were found to be increased either by treatment with the small molecule inhibitor or upon depletion of native LolCDE. Finally, we tested several antibiotics with differing mechanisms of action to identify potential specific reporter genes for the further development of compounds that would inhibit the native P. aeruginosa Lol system.IMPORTANCE A key set of lipoprotein transport components, LolCDE, were inhibited by both a small molecule as well as genetic downregulation of their expression. The data show a unique signature in the Pseudomonas aeruginosa transcriptome in response to perturbation of outer membrane biogenesis. In addition, we demonstrate a transcriptional response in key genes with marked specificity compared to several antibiotic classes with different mechanisms of action. As a result of this work, we identified genes that could be of potential use as biomarkers in a cell-based screen for novel antibiotic inhibitors of lipoprotein transport in P. aeruginosa.
Keywords: LolCDE; Pseudomonas; lipoprotein transport; molecular inhibitor; transcriptome.
Copyright © 2020 Lorenz et al.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

