Metabolic precision labeling enables selective probing of O-linked N-acetylgalactosamine glycosylation
- PMID: 32989128
- PMCID: PMC7568240
- DOI: 10.1073/pnas.2007297117
Metabolic precision labeling enables selective probing of O-linked N-acetylgalactosamine glycosylation
Abstract
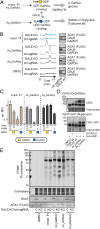
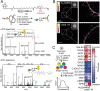
Protein glycosylation events that happen early in the secretory pathway are often dysregulated during tumorigenesis. These events can be probed, in principle, by monosaccharides with bioorthogonal tags that would ideally be specific for distinct glycan subtypes. However, metabolic interconversion into other monosaccharides drastically reduces such specificity in the living cell. Here, we use a structure-based design process to develop the monosaccharide probe N-(S)-azidopropionylgalactosamine (GalNAzMe) that is specific for cancer-relevant Ser/Thr(O)-linked N-acetylgalactosamine (GalNAc) glycosylation. By virtue of a branched N-acylamide side chain, GalNAzMe is not interconverted by epimerization to the corresponding N-acetylglucosamine analog by the epimerase N-acetylgalactosamine-4-epimerase (GALE) like conventional GalNAc-based probes. GalNAzMe enters O-GalNAc glycosylation but does not enter other major cell surface glycan types including Asn(N)-linked glycans. We transfect cells with the engineered pyrophosphorylase mut-AGX1 to biosynthesize the nucleotide-sugar donor uridine diphosphate (UDP)-GalNAzMe from a sugar-1-phosphate precursor. Tagged with a bioorthogonal azide group, GalNAzMe serves as an O-glycan-specific reporter in superresolution microscopy, chemical glycoproteomics, a genome-wide CRISPR-knockout (CRISPR-KO) screen, and imaging of intestinal organoids. Additional ectopic expression of an engineered glycosyltransferase, "bump-and-hole" (BH)-GalNAc-T2, boosts labeling in a programmable fashion by increasing incorporation of GalNAzMe into the cell surface glycoproteome. Alleviating the need for GALE-KO cells in metabolic labeling experiments, GalNAzMe is a precision tool that allows a detailed view into the biology of a major type of cancer-relevant protein glycosylation.
Keywords: bioorthogonal; glycosylation; glycosyltransferase; mucin.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
[Comparison of the performance of secretome analysis based on metabolic labeling by three unnatural sugars].Se Pu. 2021 Oct;39(10):1086-1093. doi: 10.3724/SP.J.1123.2021.04017. Se Pu. 2021. PMID: 34505430 Free PMC article. Chinese.
-
Expanding the repertoire of GalNAc analogues for cell-specific bioorthogonal tagging of glycoproteins.RSC Chem Biol. 2024 Aug 22;5(10):1002-9. doi: 10.1039/d4cb00093e. Online ahead of print. RSC Chem Biol. 2024. PMID: 39238612 Free PMC article.
-
O-GalNAc incorporation into a cluster acceptor site of three consecutive threonines. Distinct specificity of GalNAc-transferase isoforms.Eur J Biochem. 2002 Dec;269(24):6173-83. doi: 10.1046/j.1432-1033.2002.03334.x. Eur J Biochem. 2002. PMID: 12473113
-
Precision Probing of O-GalNAc Glycosylation Using Bump-and-Hole Engineering.Chimia (Aarau). 2025 Mar 26;79(3):146-151. doi: 10.2533/chimia.2025.146. Chimia (Aarau). 2025. PMID: 40156558 Review.
-
Molecular evolution and functional divergence of UDP-hexose 4-epimerases.Curr Opin Chem Biol. 2021 Apr;61:53-62. doi: 10.1016/j.cbpa.2020.09.007. Epub 2020 Nov 7. Curr Opin Chem Biol. 2021. PMID: 33171387 Review.
Cited by
-
Small molecule inhibitors of mammalian glycosylation.Matrix Biol Plus. 2022 Mar 16;16:100108. doi: 10.1016/j.mbplus.2022.100108. eCollection 2022 Dec. Matrix Biol Plus. 2022. PMID: 36467541 Free PMC article.
-
In Situ Glycan Analysis and Editing in Living Systems.JACS Au. 2024 Jan 17;4(2):384-401. doi: 10.1021/jacsau.3c00717. eCollection 2024 Feb 26. JACS Au. 2024. PMID: 38425935 Free PMC article. Review.
-
Cell-specific bioorthogonal tagging of glycoproteins.Nat Commun. 2022 Oct 25;13(1):6237. doi: 10.1038/s41467-022-33854-0. Nat Commun. 2022. PMID: 36284108 Free PMC article.
-
Newly synthesized glycoprotein profiling to identify molecular signatures of warm ischemic injury in donor lungs.Am J Physiol Lung Cell Mol Physiol. 2023 Jul 1;325(1):L30-L44. doi: 10.1152/ajplung.00412.2022. Epub 2023 May 2. Am J Physiol Lung Cell Mol Physiol. 2023. PMID: 37130807 Free PMC article.
-
Photocrosslinking and capture for the analysis of carbohydrate-dependent interactions.Bioorg Med Chem Lett. 2025 Mar 1;117:130077. doi: 10.1016/j.bmcl.2024.130077. Epub 2024 Dec 20. Bioorg Med Chem Lett. 2025. PMID: 39710139 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- FC001115/CRUK_/Cancer Research UK/United Kingdom
- R35 GM118067/GM/NIGMS NIH HHS/United States
- FC001749/MRC_/Medical Research Council/United Kingdom
- CIHR/Canada
- FC001105/CRUK_/Cancer Research UK/United Kingdom
- F32 GM126663/GM/NIGMS NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- FC001749/CRUK_/Cancer Research UK/United Kingdom
- FC001105/MRC_/Medical Research Council/United Kingdom
- R01 CA200423/CA/NCI NIH HHS/United States
- BB/F008309/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- FC001115/WT_/Wellcome Trust/United Kingdom
- 104785/B/14/Z/WT_/Wellcome Trust/United Kingdom
- HHMI/Howard Hughes Medical Institute/United States
- FC001115/MRC_/Medical Research Council/United Kingdom
- FC001749/WT_/Wellcome Trust/United Kingdom
- FC001105/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

