Nuclear receptor REVERBα is a state-dependent regulator of liver energy metabolism
- PMID: 32989157
- PMCID: PMC7568238
- DOI: 10.1073/pnas.2005330117
Nuclear receptor REVERBα is a state-dependent regulator of liver energy metabolism
Abstract
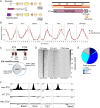
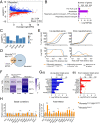
The nuclear receptor REVERBα is a core component of the circadian clock and proposed to be a dominant regulator of hepatic lipid metabolism. Using antibody-independent ChIP-sequencing of REVERBα in mouse liver, we reveal a high-confidence cistrome and define direct target genes. REVERBα-binding sites are highly enriched for consensus RORE or RevDR2 motifs and overlap with corepressor complex binding. We find no evidence for transcription factor tethering and DNA-binding domain-independent action. Moreover, hepatocyte-specific deletion of Reverbα drives only modest physiological and transcriptional dysregulation, with derepressed target gene enrichment limited to circadian processes. Thus, contrary to previous reports, hepatic REVERBα does not repress lipogenesis under basal conditions. REVERBα control of a more extensive transcriptional program is only revealed under conditions of metabolic perturbation (including mistimed feeding, which is a feature of the global Reverbα-/- mouse). Repressive action of REVERBα in the liver therefore serves to buffer against metabolic challenge, rather than drive basal rhythmicity in metabolic activity.
Keywords: NR1D1; circadian clock; energy metabolism; liver; nuclear hormone receptor.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Kervezee L., Kosmadopoulos A., Boivin D. B., Metabolic and cardiovascular consequences of shift work: The role of circadian disruption and sleep disturbances. Eur. J. Neurosci. 51, 396–412 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 107849/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- MR/P00279X/1/MRC_/Medical Research Council/United Kingdom
- 107851/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- MR/P023576/1/MRC_/Medical Research Council/United Kingdom
- BB/I018654/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/P023576/2/MRC_/Medical Research Council/United Kingdom
- MR/P011853/2/MRC_/Medical Research Council/United Kingdom
- DH_/Department of Health/United Kingdom
- MR/P011853/1 /MRC_/Medical Research Council/United Kingdom
- MR/N021479/1/MRC_/Medical Research Council/United Kingdom
- MR/N021479/1/MRC_/Medical Research Council/United Kingdom
- MR/M008908/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

