An optimized acetylcholine sensor for monitoring in vivo cholinergic activity
- PMID: 32989318
- PMCID: PMC7606762
- DOI: 10.1038/s41592-020-0953-2
An optimized acetylcholine sensor for monitoring in vivo cholinergic activity
Abstract
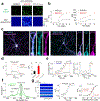
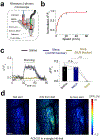
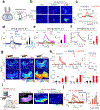
The ability to directly measure acetylcholine (ACh) release is an essential step toward understanding its physiological function. Here we optimized the GRABACh (GPCR-activation-based ACh) sensor to achieve substantially improved sensitivity in ACh detection, as well as reduced downstream coupling to intracellular pathways. The improved version of the ACh sensor retains the subsecond response kinetics, physiologically relevant affinity and precise molecular specificity for ACh of its predecessor. Using this sensor, we revealed compartmental ACh signals in the olfactory center of transgenic flies in response to external stimuli including odor and body shock. Using fiber photometry recording and two-photon imaging, our ACh sensor also enabled sensitive detection of single-trial ACh dynamics in multiple brain regions in mice performing a variety of behaviors.
Figures









References
-
- Dale HH The action of certain esters and ethers of choline, and their relation to muscarine. Journal of Pharmacology and Experimental Therapeutics 6, 147–190 (1914).
-
- Winkler J, Suhr S, Gage F, Thal L & Fisher L Essential role of neocortical acetylcholine in spatial memory. Nature 375, 484 (1995). - PubMed
-
- KrnjeviĆ K & Miledi R Acetylcholine in Mammalian Neuromuscular Transmission. Nature 182, 805–806 (1958). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

