Establishment of replication-competent vesicular stomatitis virus-based recombinant viruses suitable for SARS-CoV-2 entry and neutralization assays
- PMID: 32990161
- PMCID: PMC7594855
- DOI: 10.1080/22221751.2020.1830715
Establishment of replication-competent vesicular stomatitis virus-based recombinant viruses suitable for SARS-CoV-2 entry and neutralization assays
Abstract
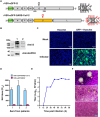
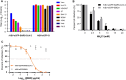
Replication-competent vesicular stomatitis virus (VSV)-based recombinant viruses are useful tools for studying emerging and highly pathogenic enveloped viruses in level 2 biosafety facilities. Here, we used a replication-competent recombinant VSVs (rVSVs) encoding the spike (S) protein of SARS-CoV-2 in place of the original G glycoprotein (rVSV-eGFP-SARS-CoV-2) to develop a high-throughput entry assay for SARS-CoV-2. The S protein was incorporated into the recovered rVSV-eGFP-SARS-CoV-2 particles, which could be neutralized by sera from convalescent COVID-19 patients. The recombinant SARS-CoV-2 also displayed entry characteristics similar to the wild type virus, such as cell tropism and pH-dependence. The neutralizing titers of antibodies and sera measured by rVSV-eGFP-SARS-CoV-2 were highly correlated with those measured by wild-type viruses or pseudoviruses. Therefore, this is a safe and convenient screening tool for SARS-CoV-2, and it may promote the development of COVID-19 vaccines and therapeutics.
Keywords: SARS-CoV-2; VSV; entry; neutralization assay; replication-competent.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
