Quantitative and time-resolved miRNA pattern of early human T cell activation
- PMID: 32990751
- PMCID: PMC7544210
- DOI: 10.1093/nar/gkaa788
Quantitative and time-resolved miRNA pattern of early human T cell activation
Abstract
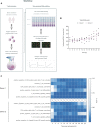
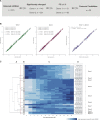
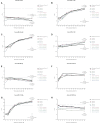
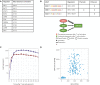
T cells are central to the immune response against various pathogens and cancer cells. Complex networks of transcriptional and post-transcriptional regulators, including microRNAs (miRNAs), coordinate the T cell activation process. Available miRNA datasets, however, do not sufficiently dissolve the dynamic changes of miRNA controlled networks upon T cell activation. Here, we established a quantitative and time-resolved expression pattern for the entire miRNome over a period of 24 h upon human T-cell activation. Based on our time-resolved datasets, we identified central miRNAs and specified common miRNA expression profiles. We found the most prominent quantitative expression changes for miR-155-5p with a range from initially 40 molecules/cell to 1600 molecules/cell upon T-cell activation. We established a comprehensive dynamic regulatory network of both the up- and downstream regulation of miR-155. Upstream, we highlight IRF4 and its complexes with SPI1 and BATF as central for the transcriptional regulation of miR-155. Downstream of miR-155-5p, we verified 17 of its target genes by the time-resolved data recorded after T cell activation. Our data provide comprehensive insights into the range of stimulus induced miRNA abundance changes and lay the ground to identify efficient points of intervention for modifying the T cell response.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

