Type II anti-CRISPR proteins as a new tool for synthetic biology
- PMID: 32991234
- PMCID: PMC8244766
- DOI: 10.1080/15476286.2020.1827803
Type II anti-CRISPR proteins as a new tool for synthetic biology
Abstract
The clustered regularly interspaced short palindromic repeats (CRISPR)-Cas (CRISPR-associated proteins) system represents, in prokaryotes, an adaptive and inheritable immune response against invading DNA. The discovery of anti-CRISPR proteins (Acrs), which are inhibitors of CRISPR-Cas, mainly encoded by phages and prophages, showed a co-evolution history between prokaryotes and phages. In the past decade, the CRISPR-Cas systems together with the corresponding Acrs have been turned into a genetic-engineering tool. Among the six types of CRISPR-Cas characterized so far, type II CRISPR-Cas system is the most popular in biotechnology. Here, we discuss about the discovery, the reported inhibitory mechanisms, and the applications in both gene editing and gene transcriptional regulation of type II Acrs. Moreover, we provide insights into future potential research and feasible applications.
Keywords: CRISPR-Cas system; Cas9; gene editing; synthetic biology; transcriptional regulation; type II anti-CRISPRs.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures
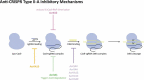

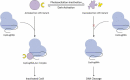

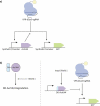
References
-
- Mojica FJ, Diez-Villasenor C, Soria E, et al. Biological significance of a family of regularly spaced repeats in the genomes of archaea, bacteria and mitochondria. Mol Microbiol. 2000;36(1):p. 244–6. - PubMed
-
- Jansen R, Embden JDAV, Gaastra W, et al. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):p. p. 1565–1575. - PubMed
-
- Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151(8):p. p. 2551–2561. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
