Translocation of Viable Gut Microbiota to Mesenteric Adipose Drives Formation of Creeping Fat in Humans
- PMID: 32991841
- PMCID: PMC7521382
- DOI: 10.1016/j.cell.2020.09.009
Translocation of Viable Gut Microbiota to Mesenteric Adipose Drives Formation of Creeping Fat in Humans
Abstract
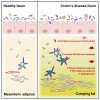
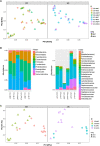
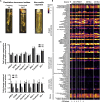
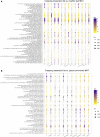
A mysterious feature of Crohn's disease (CD) is the extra-intestinal manifestation of "creeping fat" (CrF), defined as expansion of mesenteric adipose tissue around the inflamed and fibrotic intestine. In the current study, we explore whether microbial translocation in CD serves as a central cue for CrF development. We discovered a subset of mucosal-associated gut bacteria that consistently translocated and remained viable in CrF in CD ileal surgical resections, and identified Clostridium innocuum as a signature of this consortium with strain variation between mucosal and adipose isolates, suggesting preference for lipid-rich environments. Single-cell RNA sequencing characterized CrF as both pro-fibrotic and pro-adipogenic with a rich milieu of activated immune cells responding to microbial stimuli, which we confirm in gnotobiotic mice colonized with C. innocuum. Ex vivo validation of expression patterns suggests C. innocuum stimulates tissue remodeling via M2 macrophages, leading to an adipose tissue barrier that serves to prevent systemic dissemination of bacteria.
Keywords: Crohn’s disease; adipogenesis; creeping fat; fibrosis; human microbiome; ileum; inflammatory bowel diseases; macrophages; mesenteric adipose; translocation.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests S.D. and C.W.Y.H. are inventors on US patent application #62/679,624.
Figures









Comment in
-
Creeping fat in Crohn's disease explained.Nat Rev Gastroenterol Hepatol. 2020 Dec;17(12):713. doi: 10.1038/s41575-020-00379-0. Nat Rev Gastroenterol Hepatol. 2020. PMID: 33106622 No abstract available.
-
When Gut Microbiota Creep into Fat, the Fat Creeps Back.Cell. 2020 Oct 29;183(3):589-591. doi: 10.1016/j.cell.2020.10.008. Cell. 2020. PMID: 33125887 Free PMC article.
-
Creeping Fat in Crohn's Disease: Innocuous or innocuum?Immunity. 2020 Nov 17;53(5):905-907. doi: 10.1016/j.immuni.2020.10.018. Immunity. 2020. PMID: 33207215
References
-
- Amar J., Chabo C., Waget A., Klopp P., Vachoux C., Bermúdez-Humarán L.G., Smirnova N., Bergé M., Sulpice T., Lahtinen S. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol. Med. 2011;3:559–572. - PMC - PubMed
-
- Anderson M. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

