Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant
- PMID: 32991842
- PMCID: PMC7492024
- DOI: 10.1016/j.cell.2020.09.032
Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant
Abstract
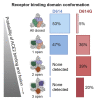
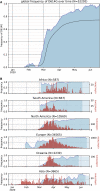
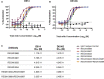
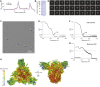
The SARS-CoV-2 spike (S) protein variant D614G supplanted the ancestral virus worldwide, reaching near fixation in a matter of months. Here we show that D614G was more infectious than the ancestral form on human lung cells, colon cells, and on cells rendered permissive by ectopic expression of human ACE2 or of ACE2 orthologs from various mammals, including Chinese rufous horseshoe bat and Malayan pangolin. D614G did not alter S protein synthesis, processing, or incorporation into SARS-CoV-2 particles, but D614G affinity for ACE2 was reduced due to a faster dissociation rate. Assessment of the S protein trimer by cryo-electron microscopy showed that D614G disrupts an interprotomer contact and that the conformation is shifted toward an ACE2 binding-competent state, which is modeled to be on pathway for virion membrane fusion with target cells. Consistent with this more open conformation, neutralization potency of antibodies targeting the S protein receptor-binding domain was not attenuated.
Keywords: ACE2; COVID-19; SARS-CoV-2; Spike protein; coronavirus; cryo-electron microscopy; infectivity; neutralizing antibody; pandemic.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests P.C.S. is a co-founder and shareholder of Sherlock Biosciences and a board member and shareholder of Danaher Corporation. J.E.L. consulted for Sherlock Biosciences. C.A.K., K.E.P., and A.B. are employed by Regeneron Pharmaceuticals and own stock options in the company. C.A.K. is an officer at Regeneron. X.W., A.B., and N.D. are employees of Thermo Fisher Scientific.
Figures





Update of
-
Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.bioRxiv [Preprint]. 2020 Jul 15:2020.07.04.187757. doi: 10.1101/2020.07.04.187757. bioRxiv. 2020. Update in: Cell. 2020 Oct 29;183(3):739-751.e8. doi: 10.1016/j.cell.2020.09.032. PMID: 32637944 Free PMC article. Updated. Preprint.
Comment in
-
Connections between biomechanics and higher infectivity: a tale of the D614G mutation in the SARS-CoV-2 spike protein.Signal Transduct Target Ther. 2021 Jan 11;6(1):11. doi: 10.1038/s41392-020-00439-6. Signal Transduct Target Ther. 2021. PMID: 33431822 Free PMC article. No abstract available.
References
-
- Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

