Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology
- PMID: 32991844
- PMCID: PMC7494283
- DOI: 10.1016/j.cell.2020.09.037
Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology
Abstract
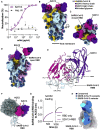
Analysis of the specificity and kinetics of neutralizing antibodies (nAbs) elicited by SARS-CoV-2 infection is crucial for understanding immune protection and identifying targets for vaccine design. In a cohort of 647 SARS-CoV-2-infected subjects, we found that both the magnitude of Ab responses to SARS-CoV-2 spike (S) and nucleoprotein and nAb titers correlate with clinical scores. The receptor-binding domain (RBD) is immunodominant and the target of 90% of the neutralizing activity present in SARS-CoV-2 immune sera. Whereas overall RBD-specific serum IgG titers waned with a half-life of 49 days, nAb titers and avidity increased over time for some individuals, consistent with affinity maturation. We structurally defined an RBD antigenic map and serologically quantified serum Abs specific for distinct RBD epitopes leading to the identification of two major receptor-binding motif antigenic sites. Our results explain the immunodominance of the receptor-binding motif and will guide the design of COVID-19 vaccines and therapeutics.
Keywords: COVID-19; SARS-CoV-2; coronaviruses; effector functions; immunity; neutralizing antibodies.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests L.P., N.C., M. Beltramello, C.S.-F., D.P., L.E.R., F.Z., N.S., J.B., A.P., S. Jaconi, B.G., A.M., A.D.M., M.S.P., E.C., S.V.G., F.J., C.H.-D., M.S., D.H., G.S., K.F., H.W.V., A.L., and D.C. are employees of Vir Biotechnology Inc. and may hold shares in Vir Biotechnology Inc. D.C. is currently listed as an inventor on multiple patent applications, which disclose the subject matter described in this manuscript. The Veesler laboratory has received a sponsored research agreement from Vir Biotechnology Inc. The other authors declare no competing interests.
Figures



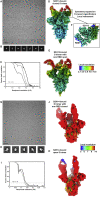



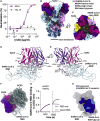
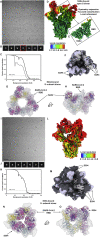
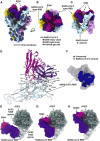

References
-
- Agirre J., Iglesias-Fernández J., Rovira C., Davies G.J., Wilson K.S., Cowtan K.D. Privateer: software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 2015;22:833–834. - PubMed
-
- Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.16. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

