Host Immune Response Driving SARS-CoV-2 Evolution
- PMID: 32992592
- PMCID: PMC7599751
- DOI: 10.3390/v12101095
Host Immune Response Driving SARS-CoV-2 Evolution
Abstract
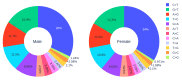
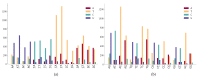
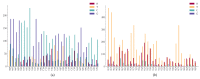
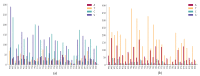
The transmission and evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are of paramount importance in controlling and combating the coronavirus disease 2019 (COVID-19) pandemic. Currently, over 15,000 SARS-CoV-2 single mutations have been recorded, which have a great impact on the development of diagnostics, vaccines, antibody therapies, and drugs. However, little is known about SARS-CoV-2's evolutionary characteristics and general trend. In this work, we present a comprehensive genotyping analysis of existing SARS-CoV-2 mutations. We reveal that host immune response via APOBEC and ADAR gene editing gives rise to near 65% of recorded mutations. Additionally, we show that children under age five and the elderly may be at high risk from COVID-19 because of their overreaction to the viral infection. Moreover, we uncover that populations of Oceania and Africa react significantly more intensively to SARS-CoV-2 infection than those of Europe and Asia, which may explain why African Americans were shown to be at increased risk of dying from COVID-19, in addition to their high risk of COVID-19 infection caused by systemic health and social inequities. Finally, our study indicates that for two viral genome sequences of the same origin, their evolution order may be determined from the ratio of mutation type, C > T over T > C.
Keywords: ADAR; APOBEC; COVID-19; SARS-CoV-2; gene editing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





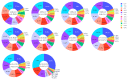

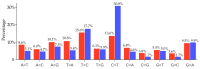
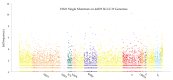
Update of
-
Host immune response driving SARS-CoV-2 evolution.ArXiv [Preprint]. 2020 Aug 20:arXiv:2008.07488v2. ArXiv. 2020. Update in: Viruses. 2020 Sep 27;12(10):E1095. doi: 10.3390/v12101095. PMID: 32839723 Free PMC article. Updated. Preprint.
References
-
- WHO . Coronavirus Disease 2019 (COVID-19) Situation Report—193. WHO; Geneva, Switzerland: 2020.
-
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K., et al. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. - DOI - PMC - PubMed
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

