Point-Of-Care or Point-Of-Need Diagnostic Tests: Time to Change Outbreak Investigation and Pathogen Detection
- PMID: 32992688
- PMCID: PMC7709694
- DOI: 10.3390/tropicalmed5040151
Point-Of-Care or Point-Of-Need Diagnostic Tests: Time to Change Outbreak Investigation and Pathogen Detection
Abstract
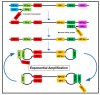
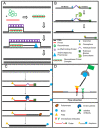
In the recent years, the progress of international trade and travel has led to an increased risk of emerging infections. Around 75 percent of the pathogens causing these infections are of animal origin. Point-of-care tests (POCT) and point-of-need tests (PONT) have been established in order to directly provide accurate and rapid diagnostics at field level, the patient bed-side or at the site of outbreaks. These assays can help physicians and decision makers to take the right action without delay. Typically, POCT and PONT rely on genomic identification of pathogens or track their immunological fingerprint. Recently, protocols for metagenomic diagnostics in the field have been developed. In this review, we give an overview of the latest developments in portable diagnostic methods. In addition, four mobile platforms for the implementation of these techniques at point-of-care and point-of-need are described. These approaches can provide reliable diagnostics and surveillance, especially in low resource settings as well as at the level of one health.
Keywords: diagnostics; field applicable diagnostics; isothermal amplification; lateral flow assay; mobile laboratory; point-of-care; point-of-need.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- World Organization for Animal Health OIE-Listed Diseases, Infections and Infestations in Force in 2019. [(accessed on 9 April 2019)]; Available online: http://www.oie.int/animal-health-in-the-world/oie-listed-diseases-2019/
-
- Gebreyes W.A., Dupouy-Camet J., Newport M.J., Oliveira C.J., Schlesinger L.S., Saif Y.M., Kariuki S., Saif L.J., Saville W., Wittum T., et al. The global one health paradigm: Challenges and opportunities for tackling infectious diseases at the human, animal, and environment interface in low-resource settings. PLoS Negl. Trop. Dis. 2014;8:e3257. doi: 10.1371/journal.pntd.0003257. - DOI - PMC - PubMed
-
- Kurpiers L.A., Schulte-Herbrüggen B., Ejotre I., Reeder D.M. Bushmeat and Emerging Infectious Diseases: Lessons from Africa. In: Angelici F.M., editor. Problematic Wildlife: A Cross-Disciplinary Approach. Springer International Publishing; Cham, Switzerland: 2016. pp. 507–551. - DOI
-
- Wilcox B.A. Forests and emerging infectious diseases of humans. Unasylva. 2006;224:11–19.
Publication types
LinkOut - more resources
Full Text Sources

