Molecular Characterization of Dengue Type 2 Outbreak in Pacific Islands Countries and Territories, 2017-2020
- PMID: 32992973
- PMCID: PMC7601490
- DOI: 10.3390/v12101081
Molecular Characterization of Dengue Type 2 Outbreak in Pacific Islands Countries and Territories, 2017-2020
Abstract
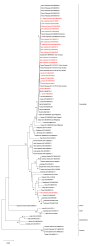
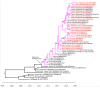
Dengue virus (DENV) serotype-2 was detected in the South Pacific region in 2014 for the first time in 15 years. In 2016-2020, DENV-2 re-emerged in French Polynesia, Vanuatu, Wallis and Futuna, and New Caledonia, co-circulating with and later replacing DENV-1. In this context, epidemiological and molecular evolution data are paramount to decipher the diffusion route of this DENV-2 in the South Pacific region. In the current work, the E gene from 23 DENV-2 serum samples collected in Vanuatu, Fiji, Wallis and Futuna, and New Caledonia was sequenced. Both maximum likelihood and Bayesian phylogenetic analyses were performed. While all DENV-2 strains sequenced belong to the Cosmopolitan genotype, phylogenetic analysis suggests at least three different DENV-2 introductions in the South Pacific between 2017 and 2020. Strains retrieved in these Pacific Islands Countries and Territories (PICTs) in 2017-2020 are phylogenetically related, with strong phylogenetic links between strains retrieved from French PICTs. These phylogenetic data substantiate epidemiological data of the DENV-2 diffusion pattern between these countries.
Keywords: Pacific; dengue; molecular evolution; phylogeny.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

