MamA essentiality in Mycobacterium smegmatis is explained by the presence of an apparent cognate restriction endonuclease
- PMID: 32993774
- PMCID: PMC7526240
- DOI: 10.1186/s13104-020-05302-z
MamA essentiality in Mycobacterium smegmatis is explained by the presence of an apparent cognate restriction endonuclease
Abstract
Objective: Restriction-Modification (R-M) systems are ubiquitous in bacteria and were considered for years as rudimentary immune systems that protect bacterial cells from foreign DNA. Currently, these R-M systems are recognized as important players in global gene expression and other cellular processes such us virulence and evolution of genomes. Here, we report the role of the unique DNA methyltransferase in Mycobacterium smegmatis, which shows a moderate degree of sequence similarity to MamA, a previously characterized methyltransferase that affects gene expression in Mycobacterium tuberculosis and is important for survival under hypoxic conditions.
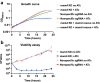
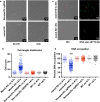
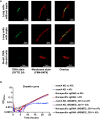
Results: We found that depletion of mamA levels impairs growth and produces elongated cell bodies. Microscopy revealed irregular septation and unevenly distributed DNA, with large areas devoid of DNA and small DNA-free cells. Deletion of MSMEG_3214, a predicted endonuclease-encoding gene co-transcribed with mamA, restored the WT growth phenotype in a mamA-depleted background. Our results suggest that the mamA-depletion phenotype can be explained by DNA cleavage by the apparent cognate restriction endonuclease MSMEG_3214. In addition, in silico analysis predicts that both MamA methyltransferase and MSMEG_3214 endonuclease recognize the same palindromic DNA sequence. We propose that MamA and MSMEG_3214 constitute a previously undescribed R-M system in M. smegmatis.
Keywords: DNA methylation; MamA; Methyltransferase; Mycobacterium smegmatis; Restriction-Modification system.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Jeltsch A. Maintenance of species identity and controlling speciation of bacteria: a new function for restriction/modification systems? Gene. 2003;317:13–16. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

