RYK-mediated filopodial pathfinding facilitates midgut elongation
- PMID: 32994164
- PMCID: PMC7648600
- DOI: 10.1242/dev.195388
RYK-mediated filopodial pathfinding facilitates midgut elongation
Abstract
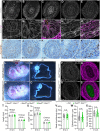
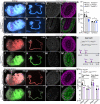
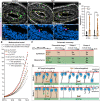
Between embryonic days 10.5 and 14.5, active proliferation drives rapid elongation of the murine midgut epithelial tube. Within this pseudostratified epithelium, nuclei synthesize DNA near the basal surface and move apically to divide. After mitosis, the majority of daughter cells extend a long, basally oriented filopodial protrusion, building a de novo path along which their nuclei can return to the basal side. WNT5A, which is secreted by surrounding mesenchymal cells, acts as a guidance cue to orchestrate this epithelial pathfinding behavior, but how this signal is received by epithelial cells is unknown. Here, we have investigated two known WNT5A receptors: ROR2 and RYK. We found that epithelial ROR2 is dispensable for midgut elongation. However, loss of Ryk phenocopies the Wnt5a-/- phenotype, perturbing post-mitotic pathfinding and leading to apoptosis. These studies reveal that the ligand-receptor pair WNT5A-RYK acts as a navigation system to instruct filopodial pathfinding, a process that is crucial for continuous cell cycling to fuel rapid midgut elongation.
Keywords: Gut elongation; Interkinetic nuclear migration; Pseudostratified; ROR2; RYK; WNT5A.
© 2020. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures




Similar articles
-
Radial WNT5A-Guided Post-mitotic Filopodial Pathfinding Is Critical for Midgut Tube Elongation.Dev Cell. 2018 Jul 16;46(2):173-188.e3. doi: 10.1016/j.devcel.2018.06.011. Dev Cell. 2018. PMID: 30016620 Free PMC article.
-
Spinal Wnt5a Plays a Key Role in Spinal Dendritic Spine Remodeling in Neuropathic and Inflammatory Pain Models and in the Proalgesic Effects of Peripheral Wnt3a.J Neurosci. 2020 Aug 26;40(35):6664-6677. doi: 10.1523/JNEUROSCI.2942-19.2020. Epub 2020 Jul 2. J Neurosci. 2020. PMID: 32616667 Free PMC article.
-
Wnt5a, Ryk and Ror2 expression in glioblastoma subgroups.Pathol Res Pract. 2015 Dec;211(12):963-72. doi: 10.1016/j.prp.2015.10.001. Epub 2015 Oct 26. Pathol Res Pract. 2015. PMID: 26596412
-
Ryk: a novel Wnt receptor regulating axon pathfinding.Int J Biochem Cell Biol. 2006;38(12):2011-7. doi: 10.1016/j.biocel.2006.07.005. Epub 2006 Aug 12. Int J Biochem Cell Biol. 2006. PMID: 16971168 Review.
-
The role of Ryk and Ror receptor tyrosine kinases in Wnt signal transduction.Cold Spring Harb Perspect Biol. 2014 Feb 1;6(2):a009175. doi: 10.1101/cshperspect.a009175. Cold Spring Harb Perspect Biol. 2014. PMID: 24370848 Free PMC article. Review.
Cited by
-
Vangl-dependent mesenchymal thinning shapes the distal lung during murine sacculation.Dev Cell. 2024 May 20;59(10):1302-1316.e5. doi: 10.1016/j.devcel.2024.03.010. Epub 2024 Apr 2. Dev Cell. 2024. PMID: 38569553 Free PMC article.
-
Human epiblast lumenogenesis: From a cell aggregate to a lumenal cyst.Semin Cell Dev Biol. 2022 Nov;131:117-123. doi: 10.1016/j.semcdb.2022.05.009. Epub 2022 May 27. Semin Cell Dev Biol. 2022. PMID: 35637065 Free PMC article. Review.
-
Spindle positioning and its impact on vertebrate tissue architecture and cell fate.Nat Rev Mol Cell Biol. 2021 Oct;22(10):691-708. doi: 10.1038/s41580-021-00384-4. Epub 2021 Jun 22. Nat Rev Mol Cell Biol. 2021. PMID: 34158639 Free PMC article. Review.
References
-
- Andre P., Wang Q., Wang N., Gao B., Schilit A., Halford M. M., Stacker S. A., Zhang X. and Yang Y. (2012). The Wnt coreceptor Ryk regulates Wnt/planar cell polarity by modulating the degradation of the core planar cell polarity component Vangl2. J. Biol. Chem. 287, 44518-44525. 10.1074/jbc.M112.414441 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

