Streamlined and Abundant Bacterioplankton Thrive in Functional Cohorts
- PMID: 32994284
- PMCID: PMC7527133
- DOI: 10.1128/mSystems.00316-20
Streamlined and Abundant Bacterioplankton Thrive in Functional Cohorts
Abstract
While fastidious microbes can be abundant and ubiquitous in their natural communities, many fail to grow axenically in laboratories due to auxotrophies or other dependencies. To overcome auxotrophies, these microbes rely on their surrounding cohort. A cohort may consist of kin (ecotypes) or more distantly related organisms (community) with the cooperation being reciprocal or nonreciprocal and expensive (Black Queen hypothesis) or costless (by-product). These metabolic partnerships (whether at single species population or community level) enable dominance by and coexistence of these lineages in nature. Here we examine the relevance of these cooperation models to explain the abundance and ubiquity of the dominant fastidious bacterioplankton of a dimictic mesotrophic freshwater lake. Using both culture-dependent (dilution mixed cultures) and culture-independent (small subunit [SSU] rRNA gene time series and environmental metagenomics) methods, we independently identified the primary cohorts of actinobacterial genera "Candidatus Planktophila" (acI-A) and "Candidatus Nanopelagicus" (acI-B) and the proteobacterial genus "Candidatus Fonsibacter" (LD12). While "Ca Planktophila" and "Ca. Fonsibacter" had no correlation in their natural habitat, they have the potential to be complementary in laboratory settings. We also investigated the bifunctional catalase-peroxidase enzyme KatG (a common good which "Ca Planktophila" is dependent upon) and its most likely providers in the lake. Further, we found that while ecotype and community cooperation combined may explain "Ca Planktophila" population abundance, the success of "Ca. Nanopelagicus" and "Ca. Fonsibacter" is better explained as a community by-product. Ecotype differentiation of "Ca. Fonsibacter" as a means of escaping predation was supported but not for overcoming auxotrophies.IMPORTANCE This study examines evolutionary and ecological relationships of three of the most ubiquitous and abundant freshwater bacterial genera: "Ca Planktophila" (acI-A), "Ca. Nanopelagicus" (acI-B), and "Ca. Fonsibacter" (LD12). Due to high abundance, these genera might have a significant influence on nutrient cycling in freshwaters worldwide, and this study adds a layer of understanding to how seemingly competing clades of bacteria can coexist by having different cooperation strategies. Our synthesis ties together network and ecological theory with empirical evidence and lays out a framework for how the functioning of populations within complex microbial communities can be studied.
Keywords: Actinobacteria; alphaproteobacteria; aquatic; bacterioplankton; common goods; ecology; evolution; metagenomics; microbial communities; networks.
Copyright © 2020 Mondav et al.
Figures




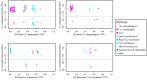

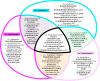
References
-
- Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, Ward DM, Rooney AP, Brambilla E, Connor N, Ratcliff RM, Nevo E, Cohan FM. 2008. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Natl Acad Sci U S A 105:2504–2509. doi:10.1073/pnas.0712205105. - DOI - PMC - PubMed
-
- Eriksson C, Forsberg C. 1992. Nutrient interactions and phytoplankton growth during the spring bloom period in Lake Erken, Sweden. Int Rev Gesamten Hydrobiol Hydrogr 77:517–551. doi:10.1002/iroh.19920770402. - DOI
LinkOut - more resources
Full Text Sources
