A Distinct Microbiome Signature in Posttreatment Lyme Disease Patients
- PMID: 32994327
- PMCID: PMC7527730
- DOI: 10.1128/mBio.02310-20
A Distinct Microbiome Signature in Posttreatment Lyme Disease Patients
Abstract
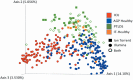
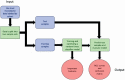
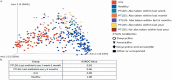
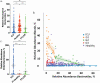
Lyme disease is the most common vector-borne disease in the United States, with an estimated incidence of 300,000 infections annually. Antibiotic intervention cures Lyme disease in the majority of cases; however, 10 to 20% of patients develop posttreatment Lyme disease syndrome (PTLDS), a debilitating condition characterized by chronic fatigue, pain, and cognitive difficulties. The underlying mechanism responsible for PTLDS symptoms, as well as a reliable diagnostic tool, has remained elusive. We reasoned that the gut microbiome may play an important role in PTLDS given that the symptoms overlap considerably with conditions in which a dysbiotic microbiome has been observed, including mood, cognition, and autoimmune disorders. Analysis of sequencing data from a rigorously curated cohort of patients with PTLDS revealed a gut microbiome signature distinct from that of healthy control subjects, as well as from that of intensive care unit (ICU) patients. Notably, microbiome sequencing data alone were indicative of PTLDS, which presents a potential, novel diagnostic tool for PTLDS.IMPORTANCE Most patients with acute Lyme disease are cured with antibiotic intervention, but 10 to 20% endure debilitating symptoms such as fatigue, neurological complications, and myalgias after treatment, a condition known as posttreatment Lyme disease syndrome (PTLDS). The etiology of PTLDS is not understood, and objective diagnostic tools are lacking. PTLDS symptoms overlap several diseases in which patients exhibit alterations in their microbiome. We found that patients with PTLDS have a distinct microbiome signature, allowing for an accurate classification of over 80% of analyzed cases. The signature is characterized by an increase in Blautia, a decrease in Bacteroides, and other changes. Importantly, this signature supports the validity of PTLDS and is the first potential biological diagnostic tool for the disease.
Keywords: Lyme disease; diagnostics; microbial communities; microflora; tick-borne pathogens.
Copyright © 2020 Morrissette et al.
Figures