Satellite glial cells promote regenerative growth in sensory neurons
- PMID: 32994417
- PMCID: PMC7524726
- DOI: 10.1038/s41467-020-18642-y
Satellite glial cells promote regenerative growth in sensory neurons
Abstract
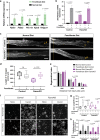
Peripheral sensory neurons regenerate their axon after nerve injury to enable functional recovery. Intrinsic mechanisms operating in sensory neurons are known to regulate nerve repair, but whether satellite glial cells (SGC), which completely envelop the neuronal soma, contribute to nerve regeneration remains unexplored. Using a single cell RNAseq approach, we reveal that SGC are distinct from Schwann cells and share similarities with astrocytes. Nerve injury elicits changes in the expression of genes related to fatty acid synthesis and peroxisome proliferator-activated receptor (PPARα) signaling. Conditional deletion of fatty acid synthase (Fasn) in SGC impairs axon regeneration. The PPARα agonist fenofibrate rescues the impaired axon regeneration in mice lacking Fasn in SGC. These results indicate that PPARα activity downstream of FASN in SGC contributes to promote axon regeneration in adult peripheral nerves and highlight that the sensory neuron and its surrounding glial coat form a functional unit that orchestrates nerve repair.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

