Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes
- PMID: 32994450
- PMCID: PMC7524749
- DOI: 10.1038/s41598-020-72264-4
Emerging MDR-Pseudomonas aeruginosa in fish commonly harbor oprL and toxA virulence genes and blaTEM, blaCTX-M, and tetA antibiotic-resistance genes
Abstract
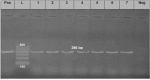
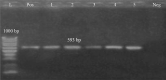
This study aimed to investigate the prevalence, antibiogram of Pseudomonas aeruginosa (P. aeruginosa), and the distribution of virulence genes (oprL, exoS, phzM, and toxA) and the antibiotic-resistance genes (blaTEM, tetA, and blaCTX-M). A total of 285 fish (165 Oreochromis niloticus and 120 Clarias gariepinus) were collected randomly from private fish farms in Ismailia Governorate, Egypt. The collected specimens were examined bacteriologically. P. aeruginosa was isolated from 90 examined fish (31.57%), and the liver was the most prominent infected organ. The antibiogram of the isolated strains was determined using a disc diffusion method, where the tested strains exhibited multi-drug resistance (MDR) to amoxicillin, cefotaxime, tetracycline, and gentamicin. The PCR results revealed that all the examined strains harbored (oprL and toxA) virulence genes, while only 22.2% were positive for the phzM gene. On the contrary, none of the tested strains were positive for the exoS gene. Concerning the distribution of the antibiotic resistance genes, the examined strains harbored blaTEM, blaCTX-M, and tetA genes with a total prevalence of 83.3%, 77.7%, and 75.6%, respectively. Experimentally infected fish with P. aeruginosa displayed high mortalities in direct proportion to the encoded virulence genes and showed similar signs of septicemia found in the naturally infected one. In conclusion, P. aeruginosa is a major pathogen of O. niloticus and C. gariepinus. oprL and toxA genes are the most predominant virulence genes associated with P. aeruginosa infection. The blaCTX-M, blaTEM, and tetA genes are the main antibiotic-resistance genes that induce resistance patterns to cefotaxime, amoxicillin, and tetracycline, highlighting MDR P. aeruginosa strains of potential public health concern.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Eissa N, El-Ghiet E, Shaheen A, Abbass A. Characterization of Pseudomonas species isolated from tilapia “Oreochromis niloticus” in Qaroun and Wadi-El-Rayan lakes, Egypt. Global Veterinaria. 2010;5:116–121.
-
- Aly, S. M. A review of fish diseases in the Egyptian aquaculture sector: Working report. (2013).
-
- Enany ME, et al. Molecular typing and evaluation of Sidr honey inhibitory effect on virulence genes of MRSA strains isolated from catfish in Egypt. Pak. J. Pharm. Sci. 2018;31:1856–1870. - PubMed
-
- El-Sayed M, Algammal A, Abouel-Atta M, Mabrok M, Emam A. Pathogenicity, genetic typing, and antibiotic sensitivity of Vibrio alginolyticus isolated from Oreochromis niloticus and Tilapia zillii. Rev. Med. Vet. 2019;170:80–86.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

