A bird's-eye view of deep learning in bioimage analysis
- PMID: 32994890
- PMCID: PMC7494605
- DOI: 10.1016/j.csbj.2020.08.003
A bird's-eye view of deep learning in bioimage analysis
Abstract
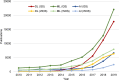
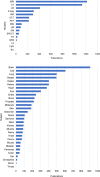
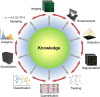
Deep learning of artificial neural networks has become the de facto standard approach to solving data analysis problems in virtually all fields of science and engineering. Also in biology and medicine, deep learning technologies are fundamentally transforming how we acquire, process, analyze, and interpret data, with potentially far-reaching consequences for healthcare. In this mini-review, we take a bird's-eye view at the past, present, and future developments of deep learning, starting from science at large, to biomedical imaging, and bioimage analysis in particular.
Keywords: Artificial neural networks; Bioimage analysis; Computer vision; Deep learning; Microscopy imaging.
© 2020 The Author(s).
Conflict of interest statement
The author declares he has no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Murphy R.F., Meijering E., Danuser G. Special issue on molecular and cellular bioimaging. IEEE Trans Image Processing. 2005;14:1233–1236. doi: 10.1109/TIP.2005.855701. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
