BioBrick-based 'Quick Gene Assembly' in vitro
- PMID: 32995504
- PMCID: PMC7513740
- DOI: 10.1093/synbio/ysx003
BioBrick-based 'Quick Gene Assembly' in vitro
Abstract
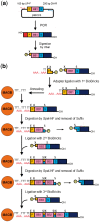
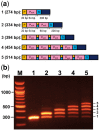
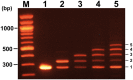
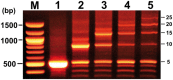
Because of the technological limitations of de novo DNA synthesis in (i) making constructs containing tandemly repeated DNA sequence units, (ii) making an unbiased DNA library containing DNA fragments with sequence multiplicity in a specific region of target genes, and (iii) replacing DNA fragments, development of efficient and reliable biochemical gene assembly methods is still anticipated. We succeeded in developing a biological standardized genetic parts that are flanked between a common upstream and downstream nucleotide sequences in an appropriate plasmid DNA vector (BioBrick)-based novel assembly method that can be used to assemble genes composed of 25 tandemly repeated BioBricks in the correct format in vitro. We named our new DNA part assembly system: 'Quick Gene Assembly (QGA)'. The time required for finishing a sequential fusion of five BioBricks is less than 24 h. We believe that the QGA method could be one of the best methods for 'gene construction based on engineering principles' at the present time, and is also a method suitable for automation in the near future.
Keywords: BioBrick; engineering principle; gene assembly; gene designing; magnetic beads.
© The Author 2017. Published by Oxford University Press.
Figures


Similar articles
-
BioBrick assembly standards and techniques and associated software tools.Methods Mol Biol. 2014;1116:1-24. doi: 10.1007/978-1-62703-764-8_1. Methods Mol Biol. 2014. PMID: 24395353
-
Methylase-assisted subcloning for high throughput BioBrick assembly.PeerJ. 2020 Sep 11;8:e9841. doi: 10.7717/peerj.9841. eCollection 2020. PeerJ. 2020. PMID: 32974095 Free PMC article.
-
In-Fusion BioBrick assembly and re-engineering.Nucleic Acids Res. 2010 May;38(8):2624-36. doi: 10.1093/nar/gkq179. Epub 2010 Apr 12. Nucleic Acids Res. 2010. PMID: 20385581 Free PMC article.
-
[Methods for construction of transgenic plant expression vector: a review].Sheng Wu Gong Cheng Xue Bao. 2015 Mar;31(3):311-27. Sheng Wu Gong Cheng Xue Bao. 2015. PMID: 26204753 Review. Chinese.
-
[Perspective on the novel methods for DNA assembly].Sheng Wu Gong Cheng Xue Bao. 2013 Aug;29(8):1113-22. Sheng Wu Gong Cheng Xue Bao. 2013. PMID: 24364348 Review. Chinese.
Cited by
-
POSoligo software for in vitro gene synthesis.Sci Rep. 2024 May 15;14(1):11117. doi: 10.1038/s41598-024-59497-3. Sci Rep. 2024. PMID: 38750104 Free PMC article.
-
Optimal parameter identification of synthetic gene networks using harmony search algorithm.PLoS One. 2019 Mar 29;14(3):e0213977. doi: 10.1371/journal.pone.0213977. eCollection 2019. PLoS One. 2019. PMID: 30925150 Free PMC article.
-
A seamless and iterative DNA assembly method named PS-Brick and its assisted metabolic engineering for threonine and 1-propanol production.Biotechnol Biofuels. 2019 Jul 15;12:180. doi: 10.1186/s13068-019-1520-x. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31338122 Free PMC article.
-
Efficient biosynthesis of D/L-alanine in the recombinant Escherichia coli BL21(DE3) by biobrick approach.Front Bioeng Biotechnol. 2024 Aug 12;12:1421167. doi: 10.3389/fbioe.2024.1421167. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39188373 Free PMC article.
-
Advancing microbiota therapeutics: the role of synthetic biology in engineering microbial communities for precision medicine.Front Bioeng Biotechnol. 2024 Dec 4;12:1511149. doi: 10.3389/fbioe.2024.1511149. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39698189 Free PMC article. Review.
References
-
- Shetty R., Lizarazo M., Rettberg R., Knight T.F. (2011) Assembly of BioBrick standard biological parts using three antibiotic assembly. Methods Enzymol., 498, 311–326. - PubMed
-
- Storch M., Casini A., Mackrow B., Fleming T., Trewhitt H., Ellis T., Baldwin G.S. (2015) BASIC: a new biopart assembly standard for idempotent cloning provides accurate, single-tier DNA assembly for synthetic biology. ACS Synth. Biol., 4, 781–787. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
