Integrated genomic view of SARS-CoV-2 in India
- PMID: 32995557
- PMCID: PMC7506191
- DOI: 10.12688/wellcomeopenres.16119.1
Integrated genomic view of SARS-CoV-2 in India
Abstract
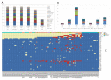
Background: India first detected SARS-CoV-2, causal agent of COVID-19 in late January 2020, imported from Wuhan, China. From March 2020 onwards, the importation of cases from countries in the rest of the world followed by seeding of local transmission triggered further outbreaks in India. Methods: We used ARTIC protocol-based tiling amplicon sequencing of SARS-CoV-2 (n=104) from different states of India using a combination of MinION and MinIT sequencing from Oxford Nanopore Technology to understand how introduction and local transmission occurred. Results: The analyses revealed multiple introductions of SARS-CoV-2 genomes, including the A2a cluster from Europe and the USA, A3 cluster from Middle East and A4 cluster (haplotype redefined) from Southeast Asia (Indonesia, Thailand and Malaysia) and Central Asia (Kyrgyzstan). The local transmission and persistence of genomes A4, A2a and A3 was also observed in the studied locations. The most prevalent genomes with patterns of variance (confined in a cluster) remain unclassified, and are here proposed as A4-clade based on its divergence within the A cluster. Conclusions: The viral haplotypes may link their persistence to geo-climatic conditions and host response. Multipronged strategies including molecular surveillance based on real-time viral genomic data is of paramount importance for a timely management of the pandemic.
Keywords: COVID; COVID-19; MinION; SARS-CoV-2; Whole Genome Sequencing; viral genomes.
Copyright: © 2020 Kumar P et al.
Conflict of interest statement
No competing interests were disclosed.
Figures


References
-
- Bhattacharyya C, Das C, Ghosh A, et al. : Global Spread of SARS-CoV-2 Subtype with Spike Protein Mutation D614G is Shaped by Human Genomic Variations that Regulate Expression of TMPRSS2 and MX1 Genes. bioRxiv. 2020. 10.1101/2020.05.04.075911 - DOI
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

