This is a preprint.
The flexibility of ACE2 in the context of SARS-CoV-2 infection
- PMID: 32995769
- PMCID: PMC7523095
- DOI: 10.1101/2020.09.16.300459
The flexibility of ACE2 in the context of SARS-CoV-2 infection
Update in
-
The flexibility of ACE2 in the context of SARS-CoV-2 infection.Biophys J. 2021 Mar 16;120(6):1072-1084. doi: 10.1016/j.bpj.2020.10.036. Epub 2020 Nov 13. Biophys J. 2021. PMID: 33189680 Free PMC article.
Abstract
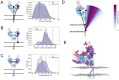
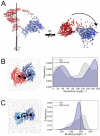
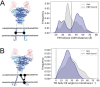
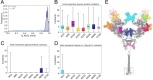
The COVID-19 pandemic has swept over the world in the past months, causing significant loss of life and consequences to human health. Although numerous drug and vaccine developments efforts are underway, many questions remain outstanding on the mechanism of SARS-CoV-2 viral association to angiotensin-converting enzyme 2 (ACE2), its main host receptor, and entry in the cell. Structural and biophysical studies indicate some degree of flexibility in the viral extracellular Spike glycoprotein and at the receptor binding domain-receptor interface, suggesting a role in infection. Here, we perform all-atom molecular dynamics simulations of the glycosylated, full-length membrane-bound ACE2 receptor, in both an apo and spike receptor binding domain (RBD) bound state, in order to probe the intrinsic dynamics of the ACE2 receptor in the context of the cell surface. A large degree of fluctuation in the full length structure is observed, indicating hinge bending motions at the linker region connecting the head to the transmembrane helix, while still not disrupting the ACE2 homodimer or ACE2-RBD interfaces. This flexibility translates into an ensemble of ACE2 homodimer conformations that could sterically accommodate binding of the spike trimer to more than one ACE2 homodimer, and suggests a mechanical contribution of the host receptor towards the large spike conformational changes required for cell fusion. This work presents further structural and functional insights into the role of ACE2 in viral infection that can be exploited for the rational design of effective SARS-CoV-2 therapeutics.
Statement of significance: As the host receptor of SARS-CoV-2, ACE2 has been the subject of extensive structural and antibody design efforts in aims to curtail COVID-19 spread. Here, we perform molecular dynamics simulations of the homodimer ACE2 full-length structure to study the dynamics of this protein in the context of the cellular membrane. The simulations evidence exceptional plasticity in the protein structure due to flexible hinge motions in the head-transmembrane domain linker region and helix mobility in the membrane, resulting in a varied ensemble of conformations distinct from the experimental structures. Our findings suggest a dynamical contribution of ACE2 to the spike glycoprotein shedding required for infection, and contribute to the question of stoichiometry of the Spike-ACE2 complex.
Figures


References
-
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., and Pöhlmann S.. 2020. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 181:271–280.e8. - PMC - PubMed
-
- Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., and Shi Z.L.. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 579:270–273. - PMC - PubMed
-
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., and Cao B.. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 395:497–506. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
