Transcriptional regulation of prolactin in a euryhaline teleost: Characterisation of gene promoters through in silico and transcriptome analyses
- PMID: 32996203
- PMCID: PMC8612711
- DOI: 10.1111/jne.12905
Transcriptional regulation of prolactin in a euryhaline teleost: Characterisation of gene promoters through in silico and transcriptome analyses
Abstract
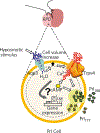
The sensitivity of prolactin (Prl) cells of the Mozambique tilapia (Oreochromis mossambicus) pituitary to variations in extracellular osmolality enables investigations into how osmoreception underlies patterns of hormone secretion. Through the actions of their main secretory products, Prl cells play a key role in supporting hydromineral balance of fishes by controlling the major osmoregulatory organs (ie, gill, intestine and kidney). The release of Prl from isolated cells of the rostral pars distalis (RPD) occurs in direct response to physiologically relevant reductions in extracellular osmolality. Although the particular signal transduction pathways that link osmotic conditions to Prl secretion have been identified, the processes that underlie hyposmotic induction of prl gene expression remain unknown. In this short review, we describe two distinct tilapia gene loci that encode Prl177 and Prl188 . From our in silico analyses of prl177 and prl188 promoter regions (approximately 1000 bp) and a transcriptome analysis of RPDs from fresh water (FW)- and seawater (SW)-acclimated tilapia, we propose a working model for how multiple transcription factors link osmoreceptive processes with adaptive patterns of prl177 and prl188 gene expression. We confirmed via RNA-sequencing and a quantitative polymerase chain reaction that multiple transcription factors emerging as predicted regulators of prl gene expression are expressed in the RPD of tilapia. In particular, gene transcripts encoding pou1f1, stat3, creb3l1, pbxip1a and stat1a were highly expressed; creb3l1, pbxip1a and stat1a were elevated in fish acclimated to SW vs FW. Combined, our in silico and transcriptome analyses set a path for resolving how adaptive patterns of Prl secretion are achieved via the integration of osmoreceptive processes with the control of prl gene transcription.
Keywords: in silico; osmoreception; prolactin; promoter; salinity; tilapia; transcription factor; transcriptome.
© 2020 British Society for Neuroendocrinology.
Conflict of interest statement
CONFLICT OF INTERESTS
The authors declare that they have no conflicts of interest.
Figures



References
-
- Bern HA, Nicoll CS. The comparative endocrinology of prolactin. Recent Prog Horm Res. 1968;24:681–720. - PubMed
-
- Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiol Rev. 2000;80:1523–1631. - PubMed
-
- Bernard V, Young J, Chanson P, Binart N. New insights in prolactin: pathological implications. Nat Rev Endocrinol. 2015;11:265–275. - PubMed
-
- Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocrin Rev. 1998;19:225–268. - PubMed
-
- Phillipps HR, Yip SH, Grattan DR. Patterns of prolactin secretion. Mol Cell Endocrinol. 2020;502:110679. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

